This web page was produced as an assignment for an undergraduate course at Davidson College.
Design and Synthesis of a Minimal Bacterial Genome

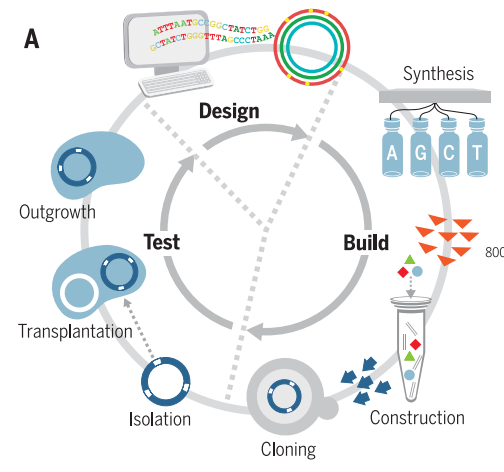
Figure 1. Process used to design, build, and test minimal genomes. Figure reproduced from Hutchinson et. al., 2016.
Project Summary and
Discovery Science:
The
genome was designed in eight segments, each of which could be tested
independently within a seven-eighths syn1.0 genome. When quasi-essential
genes were retained, a combination of redesigned segments and syn1.0
segments produced a viable genome, then refined into syn2.0. Finally,
syn2.0 was optimized to produce syn3.0, a 531 kilobase construct
containing 473 genes. Of those 473 genes, researchers could not
determine the biological function of 149, implying yet-unknown functions
that are essential to life.
This paper
demonstrates discovery science, because the researchers were not testing
a falsifiable hypothesis. They created a novel iterative
design-build-test cycle, and used this cycle to progressively minimize a
bacterial genome.
Genomic Technology:
In order
to classify genes as essential, quasi-essential, or nonessential,
researchers used Tn5
transposon mutagenesis analysis. Co-transformation of the syn1.0
genome and the Tn5 transposon produced thousands of colonies, each with
a unique transposon insertion, making up the P0 population. Insertion
sites in these cells were profiled using inverse PCR and DNA sequencing.
These cells were then serially passaged for 40 generations to select for
cells with faster growth rates, forming the P4 population. Transposon
insertion sites were profiled in both populations.
Genes with
little to no transposon insertion in either the P0 or P4 populations
were classified as essential, because any interruptions in these regions
were strongly selected against. Genes with extensive insertion in both
populations were nonessential, because the cells tolerated disruption of
these genes. Genes with insertions in the P0 generation but not the P4
generation were classified as quasi-essential, or unnecessary for life
but necessary for robust growth. Interruptions were tolerated, but not
when robust growth had been selected for.
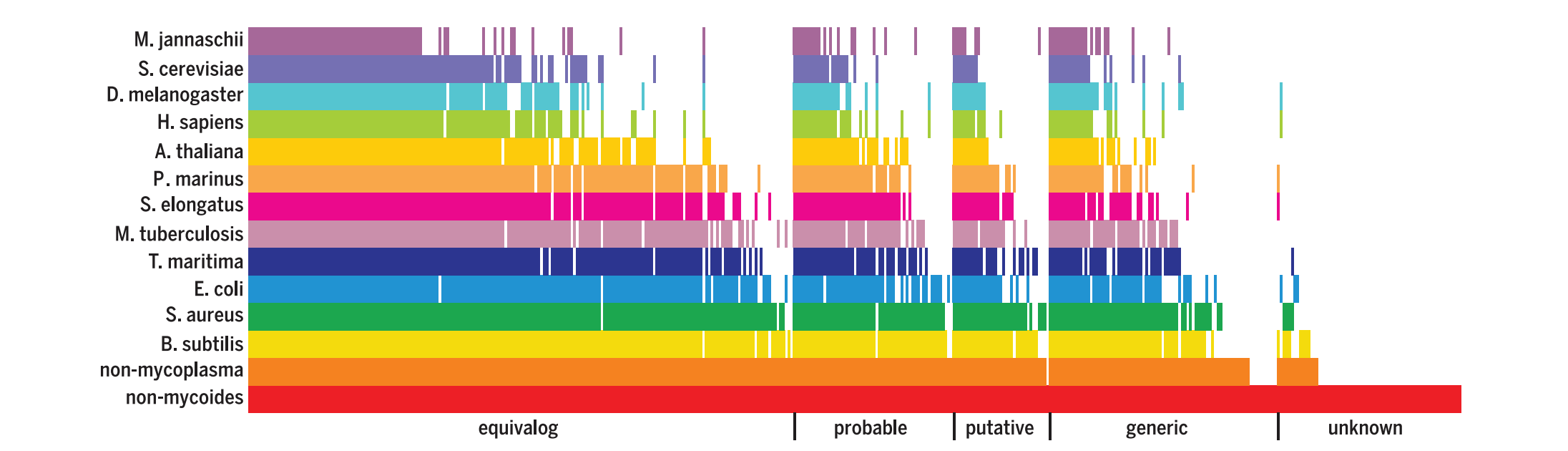
Researchers also used BLASTP alignment to analyze the similarity of syn3.0 genes to wildtype genomes of a variety of species. The visual alignment in Fig. 1 (Hutchinson et. al., 2016) shows that equivalogs of many genes retained in the minimal genome are widely represented across domains of life.
Take Home Message:
Using
transposon mutation data, researchers were able to build syn3.0, a
functional synthetic genome smaller than any that has been discovered in
nature. They retained many genes that they classified as unnecessary to
life, but minimizing the genome further would have repercussion in
growth rate. Of the 473 genes retained in the final construct, almost a
third have unknown biological functions, suggesting further research is
necessary to know more about these genes that are essential to life.
Additionally, the cyclic design-build-test model and technology to
synthesize a complete, error-free artificial genome could be used in
projects beyond genome minimization.
Evaluation:
Overall,
I found the project fascinating, specifically because of their simple
approach to a complex system. I find it surprising they ever achieved a
functioning genome, given the straightforward set of guidelines for
deletions. My impression was that genetic code, specifically non-coding
regions, is not well understood and thereby very sensitive; one wrong
base in a promoter, terminator, or other regulator sequence could render
the entire system non-functional. However, this paper implies that we
understand most of the organizational principles of genetic code, and
can delete entire non-coding segments without doing any damage.
At the
same time, it is surprising that we still cannot identify the functions
of almost a third of the essential genes. This lack of information begs
the question of what these genes are involved in. Do these proteins
perform yet-unexplored roles in processes like transcription or
translation, or are there still fundamental biological processes,
essential to life, that have not been discovered?
It makes sense that further minimization has growth-rate consequences, because it explains a natural-selection-enforced minimum on genome size. However, it does not explain why the M. mycoides genome retains non-essential genes. It seems intuitive that eliminating extraneous genes would benefit an organism by minimizing the energy required to replicate its genome. However, there must not be a very strong selection pressure, or non-essential genes would not exist in wildtype organisms. I would be interested to see an artificially-applied selection pressure or directed evolution to approach genome minimization.
References:
1. Gibson, Daniel G., John I. Glass, Carole Lartigue, Vladimir N. Noskov, Ray-Yuan Chuang, Mikkel A. Algire, Gwynedd A. Benders, et al. “Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome.” Science 329, no. 5987 (July 2, 2010): 52–56.
Genomics
Page
Biology Home Page
Hartlee's Home Page
Email Questions or Comments: hajohnston@davidson.edu
© Copyright 2017 Department of Biology, Davidson College, Davidson, NC 28035