Image courtesy of www.cnn.com
This article appears to be more of a discovery science article because the authors are not only investigating how the disease was spread, they are also showing how different tools and applications in genomics can be used for surveillance of viral epidemics. There is a hypothesis that is mentioned briefly within the results that each outbreak is the result of a different zoonotic event involving a reservoir home to many genetically diverse Ebola viruses, but I did not get the impression that this project was centered around that hypothesis at all. From this study, people can now read about what caused the Ebola outbreak of 2014 and how it was spread across the second largest continent on our planet. Hopefully other researchers are able to use the techniques applied in this study to understand other epidemics and maybe even prevent them from spreading and causing more harm. The main discovery of this article was the confirmed method of transmission and evolution of the virus during it's outbreak in western Africa as there were researchers working in close contact with patients who contracted EVD in the locations where the virus was known to be present.
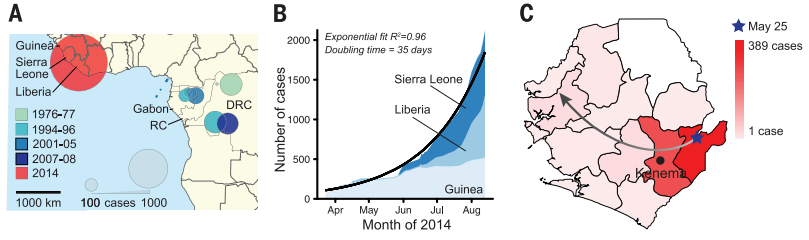
All of the utilized genomic technology was focused on analyzing the viral DNA sequence of Ebola and tracking how it has changed between different historical outbreaks and over the course of the 2014 outbreak. Two different library preparation methods and two sequencing platforms were used but Nextera library construction and Illumina sequencing was the most effective combination for comparing intrahost single-nucleotide variant (iSNV). They performed sequencing on 70% of the EVD patients in Sierra Leone in May and June. This showed many sequence differences between the 2014 Ebola virus DNA and published sequences of Ebola from past outbreaks. Additionally, they were able to find notable differences in Ebola sequences between different cases just in Sierra Leone, elucidating the diversity and fast-paced evolutionary pressures that makes the Ebola virus a such a dreaded pathogen. BEAST dating was used to construct an accurate phylogenetic tree depicting the timeline of the Ebola virus over the course of the many published outbreaks. BEAST dating uses bayesian molecular clock dating to track the amount of change in the genetic sequence of the Ebola virus to produce the most likely divergences between strains of the virus (dos Reis, 2016).
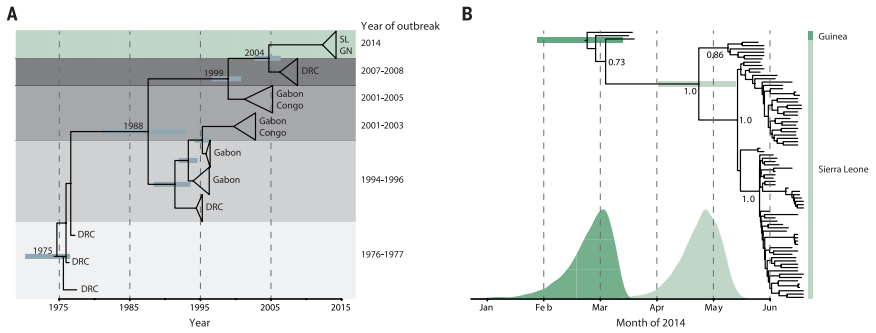
Figure 2. Phylogenetic trees produced from sequencing and BEAST dating of Ebola DNA from outbreaks over the last five decades (A) and within the 2014 outbreak (B).
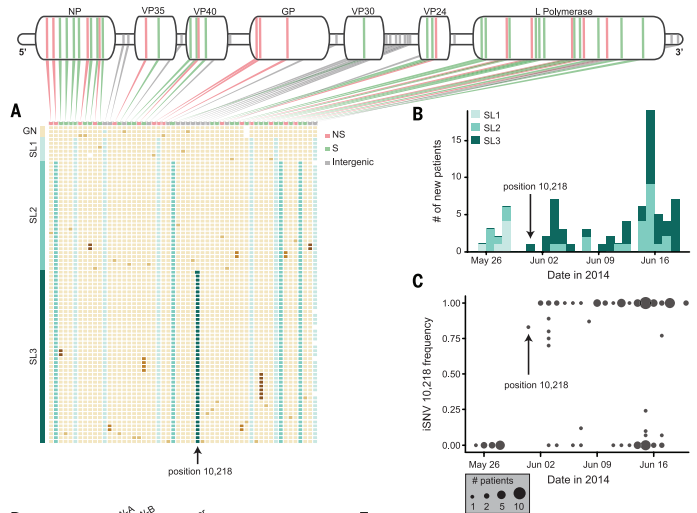
Figure 3. Mutations to the Ebola genome produce variation within the outbreak specifically in Sierra Leone. Each patient's Ebola sample is represented by one row with the corresponding mutations (A). There were different types of the virus seen throughout the outbreak, originating with SL1 and SL2 and ending predominantly with SL2 and SL3 (B). iSNV frequency shows the appearance of a novel SNP in late May and it's confirmed presence in sample from the remainder of the outbreak (C). The mutation causing the formation of SL3 is found at position 10,218.
The Ebola virus is able to quickly accumulate mutations and interact with the environment in a way that allows a fast-paced evolution and transmission leading to a serious epidemic. Understanding the dynamics of this virus during an outbreak can help prevent future outbreaks and/or facilitate an appropriate response to alleviate and epidemic caused by a virus similar to Ebola. Analyzing an epidemic with the genomic tools used in this project can uncover the story behind a specific outbreak in terms of it's origination, transmission, evolution, and location at certain points in time. However, tracking an epidemic while in the midst of it's transmission is very dangerous. In fact, five co-authors of this project passed away during this study after they contracted EVD. In most places around the world, society is packed into urban living situations where there is inevitable contact with others on a daily basis. This presents major issues for a potential serious epidemic introduced to a major city because of the magnified rate of disease transmission urban areas as stated by the article.
While this project did not directly aid those affected by the 2014 epidemic, it has provided valuable information on many aspects of the outbreak that could prove to be useful in handling future viral outbreaks in a large population. One thing that the article does not address is if the genetic differences between different populations of the virus is related to the severity of EVD. A study investigating the severity of the disease with defined genetic differences between strains could present strong data for targeting certain parts of the Ebola genome in attempt to fight against future outbreaks. This article provided a great foundation for understanding the genetic aspect of a virus and how there is variety in it's genome over time and in certain geographic locations. I am sure that there are more steps that can be taken to comprehend specific points of a specific virus' genome to potentially design drugs, vaccines, or therapies to combat the nasty symptoms presented by deadly pathogens like Ebola.
Reference List
Gire
SK, Goba A, Andersen KG, Sealfon
RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, et al.
2014.
Genomic surveillance elucidates Ebola virus origin and transmission
during the
2014 outbreak. Science 345:1369–1372.
http://science.sciencemag.org/content/345/6202/1369/tab-pdf
dos
Reis
M, Donoghue PCJ, Yang Z. 2016. Bayesian molecular clock dating of
species
divergences in the genomics era. Nat. Rev. Genet. 17:71–80.