

Quinoa genome could see 'super-food' prices tumble
Growing quinoa plant, and quinoa used for cooking: Images borrowed from: McGrath, 2017 ; DETOXINISTA
Chenopodium quinoa
(quinoa) is a south American harvested plant with the common descriptors
of "super food" and "mother grain". This crop has increased in
international demand over the last 10 years due to its nutritious and
gluten-free properties. Quinoa primarily grows in high altitude and cool
temperatures which limits possible land for crop growth. The increased
demand and stagnant supply has contributed to quinoa's heightened price
(McGrath, 2017).
A recently published sequence of the quinoa genome could lead to a solution for the shortage of the plant (Jarvis et al.,2017). Once genes that can beneficially alter quinoa are identified, quinoa can be breed for traits that optimize production rates. For example, shorter stockier plants could boost production by decreasing the amount fallen over plants. Scientist hope modifications to the quinoa genome could lead to crop production in a variety of soils, thus increasing world wide farming areas for quinoa. Modifications to the quinoa genome could lead to an increased supply that could decrease quinoa's price as low as that of wheat (McGrath, 2017).
Using current genomics
tools, Jarvis et al., developed a chromosome-scale reference
genome for quinoa (2017). Additionally, two ancestral diploid versions
modern quinoa and multiple tetraploid species were sequenced to further
identify quinoa sub-genomes (Figure 1) and to find preserved regions in
the genome with desirable traits. Quinoa orthologues to closely related
species lead to identification of 97.3% of quinoa genes. One
of the first genes to be identified was a transcription factor that
could be involved in saponin formation. Saponin is a bitter tasting
compound that quinoa uses as an environmental defense. From this
project, we have at least one gene that can be reduced or eliminated to
make quinoa more profitable by reducing treatment costs to remove the
compound.

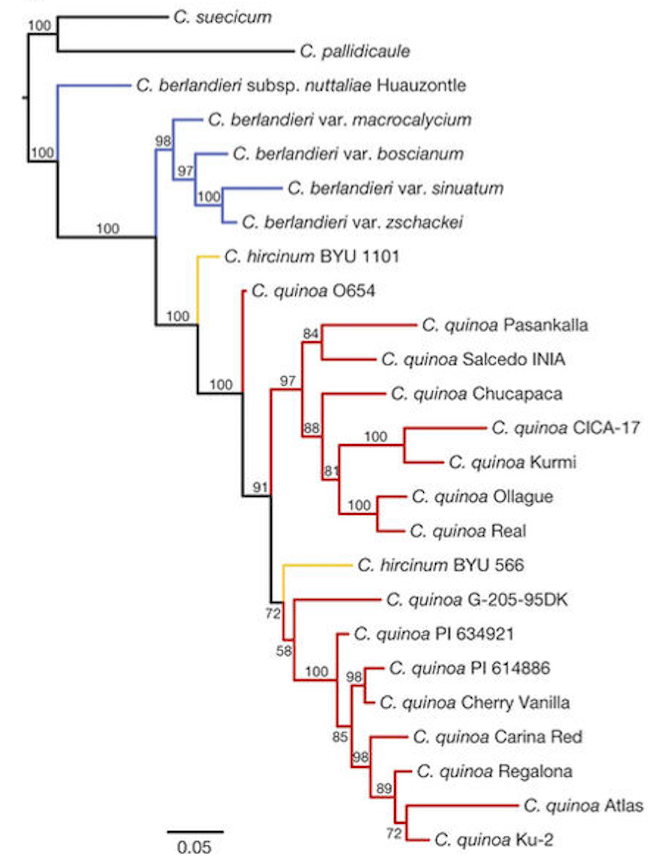
Figure 1. Multiple genomes were sequenced, including 3 species in the Chenopodium
genus, to construct a philogenetic tree of quinoa.
The quinoa project can be
classified as discovery science since there was no established
testable hypothesis. Jarvis et al, primarily focused on
sequencing the quinoa genome and identifying quinoa genes that could be
alternated for optimized crop production.
A wide array of genomics
technologies were utilized for the successful sequencing of quinoa.
First, the coastal Chilean quinoa fragmented genome was sequenced using
single-molecule real-time (SMRT) sequencing from Pacific Biosciencies
(PacBio). The sequenced genome was computationally assembled into
scaffolds using chromosome-constant and optical maps. Structures similar
to chromosomes called psuedomolecules were assembled from scaffolds
using linkage maps. RNA-seq and PacBio isoform were used to predict
protein-coding and microRNA genes. Linkage mapping and bulk segregant
analysis were used to find a single gene that controls seed saponins as
well as other correlated genes.
Through a vast number of
genomics technologies, the majority of the quinoa genome has been
sequenced. Nine hundred and fifty six quinoa genes have been identified
and annotated. With the improved sequencing of the quinoa genome, genes
such as one predicted to control sapoin production, can be selected
against by humans to decrease the processing costs to prepare quinoa for
distribution. Other genes can be selected for to improve the production
rate of the plant that can ultimately increase the supply and decrease
the cost of the internationally in demand quinoa plant.
Overall, I am impressed by
the techniques used in this project as well as the possible
international impacts. This project is very thorough. Jarvis et al.,
commonly used multiple genomics technologies for one experiment. For
example, when assembling the quinoa genome after sequencing, researchers
used both optical and chromosome-contact maps. Optical maps from BioNano
provide genetic information about molecular organization.
Chromosome-contact maps look at overall conformation of chromosomes.
When determining gene function, researchers looked at both ancestral
forms or quinoa as well as closely related species. By looking at the
same quinoa genome using multiple technologies as well as multiple
references, findings seem more compelling that if only one technology
was used.
This project was successful
in identifying and annotating 956 (97.3%) genes in the quinoa genome.
However I am curious about the sequencing or annotating limitations that
explain why 2.7% of the genes are still not identified. I am also still
curious about previous limitations that only allowed for partial quinoa
genome sequencing.
Due to the primarily
sequencing nature of this project, and because of the multiple
technologies used throughout the project, Jarvis
et al., were able to provide their results in a very compelling
way. However, they identified potential genes primarily by comparing
sequences of orthologue genes of closely related species. However, by
using primarily orthologue sequences, we may not detect genes novel to
quinoa.
The work by Jarvis et al.,
is very exciting. Many people across the world have begun to eat quinoa,
this finding allows for the possibility of greater production rates of
the coveted plant. Often times, scientific discoveries are amazing, but
there's not a large audience to applaud them. Quinoa has a large
consumer market that could be thrilled by the news. Additionally this
project has the potential to lead to direct job creation, economic
improvement and could ultimately give access to a cheaper nutritional
food source for countries in desperate need. Although a small percentage
of the quinoa genome still needs to be annotated, knowing most of the
quinoa genome establishes the possibility for future beneficial genome
modifications.
Sources
Jarvis DE et al.,
2017. The genome of Chenopodium quinoa. Nature
542:307-312. http://dx.doi.org/10.1038/nature21370
McGrath M. 2017. Quinoa genome could see 'super-food' prices tumble. BBC science and environment; [cited 208 Jan 27]. Available from: http://www.bbc.com/news/science-environment-38908321
Genomics
Page
Biology Home Page
Email Questions or Comments: itcuellar@davidson.edu
© Copyright 2018 Department of Biology, Davidson College, Davidson, NC 28035