
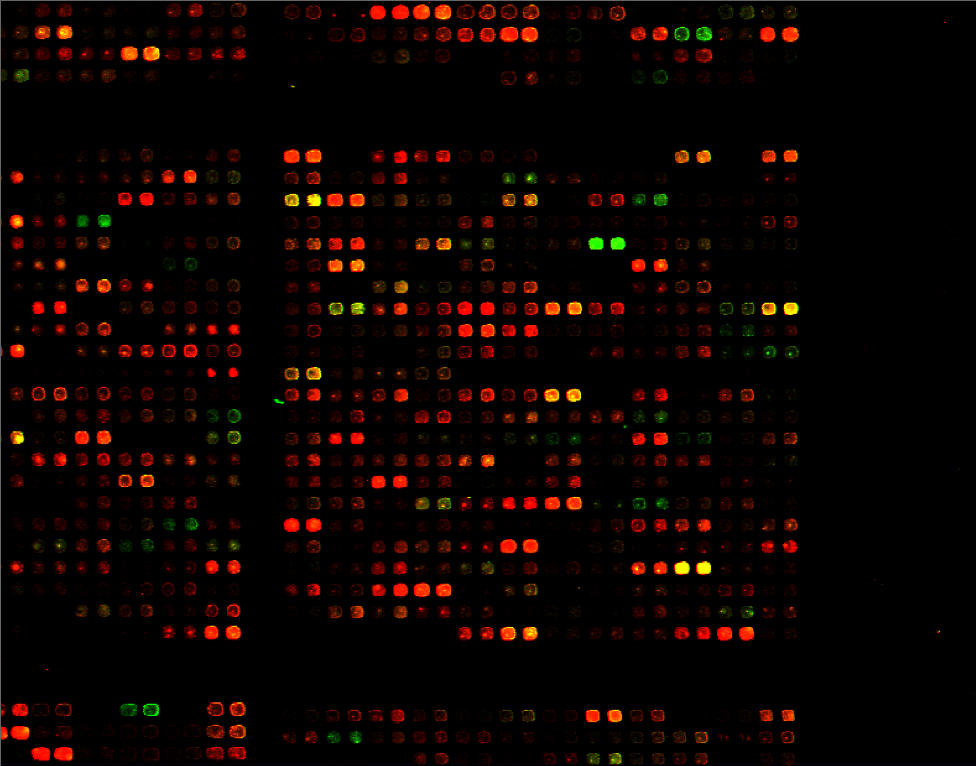
 Click
to see a larger version of this image.
Click
to see a larger version of this image. 2008 GCAT Microarray Workshops
Two Sessions; 20 people per session
Week of July 13 - 19
California State University, Fullerton
2008 Workshop #1 (20 participants)
2008 Workshop #2 (20 participants)

2008 Instructors
Todd Eckdahl, Laurie Heyer, Anne Rosenwald,
Consuelo Alvarez,
Charles Hauser, Malcolm Campbell, Edison Fowlks
Below please find some links for directions and a campus
map—(on the
map it is building H for Humanities/ Social Sciences, note location of McCarthy
Hall [MH] and Dan Black Hall [DBH] where the computer room [MH501] and experimental
labs [DBH 104 and 161] for the workshop are located). Let me know about the
number of parking passes you will need. Looking forward to the workshop!
Pre-Workshop Reading and Problem Set |
|---|
Highlights:
Free Software and Training
Interdisciplinary Team Participation Encouraged
Produce RNA; cDNA probes; Hybe; Wash; Scan; Analyze
Yeast full-genome microarrays
Computers Provided but You Can Bring Laptops
The first change is the organization of the days, as seen above. Part of this schedule adjustment is required because of the second major change, the use of Genisphere’s 3DNA method instead of direct incorporation. Finally, we will no longer rely on burning CDs and will purchase GCAT Flash Drives (1 – 2 GB each) for the participants (with space for initials). This will allow us to deliver the workshop materials and for them to archive all their data. We will collect all the drives during the scanning period and return them with all the tiff files the next morning in time for dry lab.
Workshop #2 will spend the afternoon of Day 2 in the wet lab. At this time, they will freeze their cDNA. At the end of Day 3, workshop #2 will spend about 1.5 hours in the wet lab starting the overnight hybe. At this same time, workshop #1 will be finished in the lab and they will move to a classroom for curriculum discussions.
Another major change is that participants will prep their own RNA. We will provide for them frozen yeast pellets and the RNA isolation kits. They will analyze the quality of their RNA by spectrophotometer only (i.e., will not run gels) and decide if they want to use their RNA or some prepared by the instructors prior to the workshop. Participant RNA isolation will take place on Day 2 for both workshops during their half day of wet lab.
Selection Criteria
Maximum Impact on African American, Hispanic, Native American, and Pacific Islander Students
Previous Experience with Molecular Methods (e.g., pipetting, etc.)
OR
Mathematics/Computer Science Faculty (Math/CS + Biology teams welcomed)
Interested in Bringing Microarrays into Undergraduate Curriculum
Minimal Experience with Microarrays Preferred
Willingness to Learn and Collaborate with Other Faculty
Applied to previous Workshop but Not Accepted
(please include this information on your application)
Time of Application Submission
(used as tie breaker)
GCAT does not set quotas; we try to accomodate all qualified applicants
Purpose: Genome Consortium for Active Teaching (GCAT) offers hands-on workshops for faculty who conduct research with undergraduate students and/or teach undergraduate courses to learn about gene expression analysis via microarrays.
Workshop Organization:
Part 1 of the workshop
begins with data
analysis (see schedule below)
to introduce the microarray method and will cover data analysis using
public domain data from the literature and from GCAT. Participants
will learn public domain and open source MAGIC
Tool spot-finding and analysis software. The MIAME background
information and its use in microarray evaluation will be covered.
Participants will analyze data sets in short projects. Each of the
two workshops has room for 20 participants.
Part 2 will be a hybridization workshop which will involve hands-on preparation of fluorescently labeled probes for yeast expression microarrays, their hybridization, data acquisition and data analysis using the methods presented in part 1 of the workshop.
Part 3 concludes with data analysis of the microarrays you produced. This will also serve to reinforce what you learned about data analysis.
Where: California State University, Fullerton
When: workshop #1: July 14 - 18; and workshop #2: 15 - 19, 2008
Costs and Scholarships: If funded by the NSF, this workshop will be completely free (housing, food, lab fees) for each participant with one catch. You pay for your airfare in advance but you will be reimbursed if you submit an action plan for implementing microarrays in your curriculum and submit receipts. For those with financial need for airfare in advance, you may petition to have this paid in advance.
Download Workshop Announcement |
Download Workshop Application |
|---|---|
Announcement
in PDF format (.pdf) |
Application
in PDF format (.pdf) |
Announcement
in RTF format (.rtf) |
Application
in RTF format (.rtf) |
Announcement
in Word format (.doc) |
Application
in Word format (.doc) |
Email Edison Fowlks or Malcolm Campbell with Questions
See photos from 2003 workshops
See photos from 2004 workshops
See photos from 2005 workshops
See photos from 2007 workshops
This material is based upon work supported by the National Science Foundation under Grant No. DBI-0627478. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
© Copyright 2008 Department of Biology, Davidson College,
Davidson, NC 28035
Send comments, questions, and suggestions to: macampbell@davidson.edu