Largest Open Reading Frame
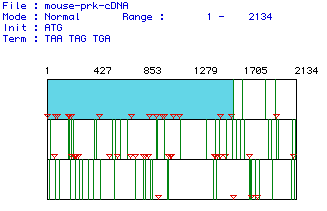
The cDNA encoding for pyruvate kinase was analyzed using MacDNAsis, and the following information was obtained.
Largest Open Reading Frame

You can find the DNA sequences that code for pyruvate kinase for five organisms by clicking here.
When translated into its predicted amino acid sequence, Mus musculus
pyruvate kinase has a predicted molecular weight of 58.01 kDa.
See the predicted amino acid content of the enzyme by clicking here.
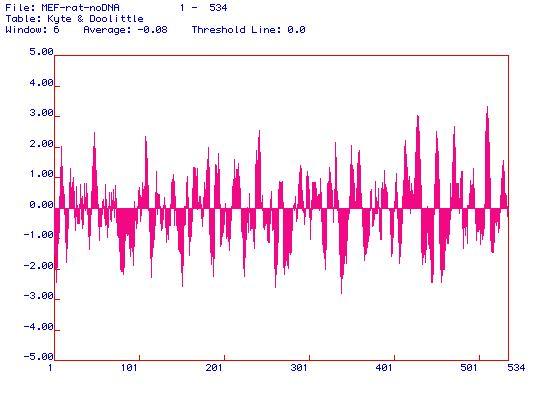
For Kyte-Doolittle plots, possible membrane-spanning domains are indicated by values above 1.8. This enzyme has many such instances, including (approximately) those close to residues 10, 60, 105, 240, 410, 440, 460, 480, and 515. Apparently, pyruvate kinase is quite membrane-associated, as it has multiple membrane-spanning domains.
To learn a little more about Kyte & Doolittle, see this page by Biotools Incorporated.
For Hopp-Woods plots, possible antigenic areas are those that are most positive, indicating the greatest degree of hyrophilicity. These areas are the epitopes most likely to bind to antibodies. This graph can help determine which portion of the enzyme to use in order to generate a peptide for synthesis of a monoclonal antibody against the peptide. This antibody will recognize a linear epitope while the enzyme is in its native conformation. The best possible candidates are areas near residues 160, 180, and 230, although many other areas (indicated by positive spikes in the graph) may possibly be antigenic. For this enzyme, I would generate a peptide from residues 175-195, because that spike seems to be the most hydrophilic. Another option would be to use residues 340-360 because --although not as high as the spike near residue 180-- that is the broadest hydrophilic region of the protein.
For a summary on Hopp & Woods, check out this page from Biotools Incorporated.

The sequences of five organisms were analyzed and compared to find
similarities. Of the five organisms, three were very similar in sequence,
while the other two were less similar. The organisms analyzed were
Saccharomyces
cerevisiae, Oryctolagus cuniculus,
Mus
musculus,
Homo sapiens,
and Chlamydia trachomatis.
Click on the species to see their amino acid sequences in a text format.
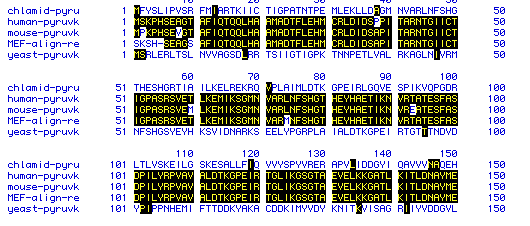
It is apparent from the figure that the highest degree of similarity is among the mammalian pyruvate kinase amino acid sequences. The protozoan and yeast protein sequences are not very similar in sequence.
In order to examine the degree of amino acid conservation over time,
a phylogenetic tree was constructed using all 5 proteins, from human,
mouse,
rabbit,
yeast,
and protozoa. (click to see
the amino acid sequences)

Figure 6: A phylogenetic tree showing the genetic
distance between each of the sequences, from top to bottom: human,
mouse, rabbit, yeast, and protozoa.
From this data, it is apparent that the genetic distance between mammals is very short; they diverged very recently along the evolutionary timeline. On the other hand, yeast and protozoa seem to have diverged much longer ago. These results are not surprising, as they are very consistent with modern theories of evolution.
Back to my main page.
Back to Davidson College Molecular
Biology Homepage.
email me at erferguson@davidson.edu