|
DNA Microarrays
|
|||||||||||
|
This page was created as an assignment for an undergraduate class at Davidson College |
|||||||||||
|
DNA microarrays are a new molecular tool with the potential to revolutionize
diagnostics, gene identification, and the study of gene expression patterns.
The basis of microarrays is a chip, similar to a computer microchip,
only it is labeled with oligonucleotides that are capable of binding
to complementary strands of DNA. In the past genes could only be examined
one at a time. DNA chips, however, allow researchers to analyze huge
numbers of genes all at once. The ability to examine patterns of gene
expression allows much greater insight into the function of gene pathways
instead of just the function of single genes.
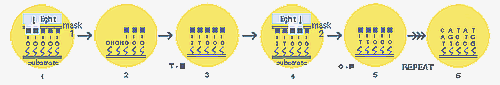
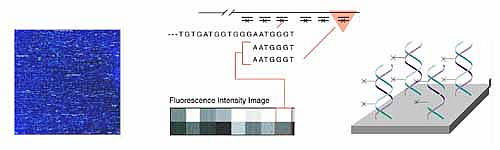
Other companies have developed variations on the Affymetrix chips. Hyseq, Inc. developed chips tagged with fluorescently labeled known oligos. When these are mixed with an unknown DNA sample, a fluorescent spot appears everywhere that the DNA binds, and the sequence of that DNA can be detected based on the known oligo sequence to which it bound. A computer is used to piece together the partial DNA sequences after the binding process is repeated several times. Incyte Pharmaceuticals has created a glass chip that has an array of gene fragments that are much longer (500 to 5000 base pairs). Messenger RNA can be isolated from two tissue samples, for example, one normal sample and one diseased sample or one sample treated with a drug. Each RNA sample is labeled with a different color fluorescent tag and then mixed with the same chip. When the chip is then scanned for the two different colors, the gene expression patterns of each sample can be compared.
Nanogen researchers have discovered a way to speed up the time required to allow the oligos to bind to their complementary DNA. The known oligos are maneuvered on the chip using an electric field. This allows the oligos to "go out and get" their complementary DNA rather than "wait" passively for it to float by and bind.
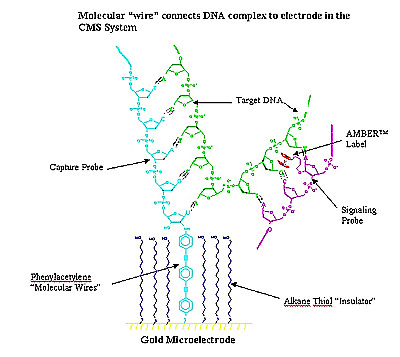
Another electronic method is being used by Clinical Microsensors, who have abandoned the use of fluorescent tags for detecting the location of DNA fragments on the chips. They use a separate probe that carries iron and will attach to DNA that has bound to its complementary oligo on the chip. The oligos are attached on a grid of electrodes that can detect the presence of the iron and indicates the position of the bound DNA.
DNA microarrays have countless applications in the world of genomics, diagnostics, and pharmaceuticals. These chips have been used for diverse purposes in recent research. DNA microarrays were used to compare different strains of bacteria that cause tuberculosis in order to better understand the evolution of these bacterial strains, an understanding that will lead to better preventative and diagnostic care for tuberculosis (Behr et al., 1999). Iyer et al. (1999) used DNA microarrays to give a temporal account of human fibroblast activity at the level of gene expression in response to fetal bovine serum (FBS), a provider of growth factors, and suggest that fibroblasts play an even larger role in the physiology of wound repair than was previously thought. A study of the genome of Saccharomyces cerevisiae using DNA microarrays allowed researchers to scan, map, and analyze allelic variation between two strains of yeast without performing time consuming tasks such as PCR or creating new DNA constructs (Winzeler et al., 1998). In the future, DNA chips could be used to make commonplace diagnostic techniques such as diagnosing strep throat even cheaper, faster, and more accurate. In the meantime, these nine square centimeter chips have already lead to the discover of PLA2, a protein produced by prostate cancer cells (and not healthy prostate cells), a protein called melastatin, which is produced by melanoma cancer cells, and mutations in BRCA1, the familiar breast cancer gene. |