 Figure
1.
Figure
1.CLONTECH'S NEW
pAP1-SEAP, pSRE-SEAP, pGRE-SEAP Vectors
As advertised in the
October 1998 issue of CLONTECHniques,
CLONTECH's new reporter vectors might be able
to improve how researchers study the activation of DNA sequences in each
vector. According to the advertisement, these vectors give researchers
the ability to "quantify transcriptional activity" and to study how the
enhancers AP1 (activator protein 1), SRE (serum response enhancer
element), or GRE (glucocorticoid response element),
when bound to the basal promotor of the herpes simplex virus (PHSV-TK),
help to induce transcription of the SEAP reporter gene. This may
lead to a better understanding of how intracellular proteins and extracellular
serum or growth factors
affect certain signal transduction pathways induced by these enhancers.
SEAP VECTORS
Each of these
new vectors contain the secreted alkaline phosphatase (SEAP) reporter gene,
so
transcriptional activity can be quantitatively measured using
CLONTECH's Great
Escape
SEAP Chemiluminescence Detection Kit.
The SEAP enzyme is secreted into
the culture medium at the same rate that intracellular concentration of SEAP
mRNA and protein change. By collecting samples at various time points,
researchers can measure the transcriptional activity of the cell without lysing
it. Therefore, there
is no need to prepare cell lysates to assay for the reporter protein, a procedure
that has been required
in the past. Also, because the kinetics of gene expression are being studied
by sampling the medium repeatedly, set-up variability is eliminated. Both
SEAP and luciferase, present in fireflies, are "highly sensitive enzymatic
reporter genes that can provide quantitative data on induction levels" (1).
pAP1-SEAP, pSRE-SEAP, and pGRE-SEAP Signal Transduction Pathways
pAP1-SEAP is used for monitoring
the induction of the activator protein 1 (AP1) and the stress-
activated protein kinase/Jun N-terminal
kinase (SAPK/JNK) signal transduction pathway (1).
This pathway can be induced by
serum and growth factors. These stimuli cause
the cellular transcription factors Jun and Fos to bind to the AP1 sequence
binding site, resulting in activation of transcription. When
these proteins are continuously expressed, cells can
grow uncontrollably, eventually resulting in tumor development (1).
Investigation of transcriptional activity from AP1
may help researchers to develop a cure for cancer by discovering how to
inhibit continuous expression of these proteins.
pSRE-SEAP is used for monitoring the induction of
SRE and the mitogen activated protein kinase (MAK) signal transduction
pathway. pGRE-SEAP is used for monitoring the induction of GRE and
the glucocorticoid mediated signaling transduction pathway.
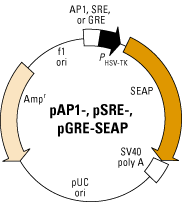
Figure 1 (1) below shows a combined vector map of pAP1-SEAP, pSRE-SEAP, and pGRE-SEAP. AP1, SRE, and GRE are the enhancers located upstream of the SEAP gene and, in the presence of serum or growth factors, bind to PHSV-TK. The SEAP coding sequence is followed by the SV40 polyadenylation signal to make sure that efficient processing of the SEAP mRNA transcript occurs. The vector also contains an f1 origin of replication for single-stranded DNA production, a pUC origin of replication, and an ampicillin resistance gene. These are beneficial for propagation and selection in E. coli (2).
 Figure
1.
Figure
1.pTK-SEAP and pTK-Luc (luciferase)
are used as negative controls to study how well enhancers
induce transcription because of
their low basal transcriptional activity. The pTK-Luc contains the
gene encoding firefly luciferase as
its reporter.
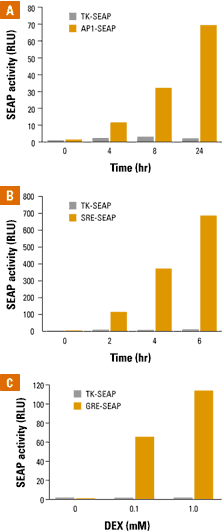
Figure 2 (1) shown below depicts data from kinetic time-course and dose-response studies using the enhancerless vector pTK-SEAP. These studies show quantitative induction levels of SEAP serum was added to cells transfected with pAP1-SEAP, pSRE-SEAP, and pGRE-SEAP.
Panel A shows a kinetic time-course study
in which human embryonic kidney (HEK 293) cells were transfected with
pAP1-SEAP. Serum was added to the medium and samples were assayed
at 0, 4, 8, and 24 hours using the
Great EscAPE Chemiluminescent Assay. It is evident that an increasing
amount of SEAP was secreted from the
cell over a 24 hour time span.
Panel B shows a kinetic
time-course study in which HEK 293 cells were transfected with pSRE-SEAP.
Serum was added and samples were assayed at
0, 2, 4, and 6 hours. It is evident that the enhancer SRE induces
transcription at a much faster rate than AP1.
Panel C shows a
dose-response study in which Saos-2 cells were transfected with pGRE-SEAP
and were incubated
overnight in a serum-free medium. Dexamethasone
(DEX) was added at a concentration of 0, 0.1, and 1.0 mM for
24 hours and SEAP activity was measured after
24 hours. Data shows that the GRE induces transcriptional activity
of SEAP.
Figure 2.
1. CLONTECHniques 1998 October. New
reporter and expression vectors: Quickly investigate
key signal transduction
pathways. <http:www.clontech.com/archive/OCT98UPD/MamTrans.html>.
Accessed 1999 Feb 2.
2. CLONTECH. 1998 October. pAP1-, pSRE-, and pGRE-SEAP
Vector Information.
<http://www.clontech.com/clontech/Vectors/pAP1-SEAP.html>.
Accessed 1999 Feb 2.
Copyright 2000 Department of Biology, Davidson College,
Davidson, NC 28036
Please send any questions, comments, or concerns to
ambradford@davidson.edu