GPS-1 Genome Priming System
The new GPS-1 Genome Priming System allows researchers to prepare
DNA segments for sequencing faster than any other method available today.
With the race to complete the Human Genome Project heating up, this new
system will allow scientists to pick up the pace. Furthermore sequencing
is the best method of learning a proteinís amino acid sequence,
so this exciting new method will enhance molecular biologyís understanding
of protein conformation by allowing them to study more sequences in less
time. During this golden age of molecular biology, the GPS-1 Genome
Priming System fits right in with the other amazing advances of the day.
How Does It Work?
The system uses Tn-7 transposons (Transprimer) (1) that randomly
insert into target DNA with universal primers. Transposons are segments
of DNA that can move to differents spots along a chromosome. Therefore,
they can move anywhere along a genome. When this in vitro method
is carried out with billions of molecules, it can be assured that the transposons
will insert in thousands of different sites within the Genomic Library.
Once the transposon has been added to a genomic library, the primers allow
template strands of the different fragments to be produced. These
strands can then be sequenced using any method available and overlaps can
be compared to discover the DNA sequence. Because the transposon
is inserted randomly, an entire genome can be sequenced at once, as opposed
to cumbersome, time-consuming methods like primer walking. Also new
primers do not need to be developed because the transposon comes with primers
ready to be used.
Figure 1. (1)
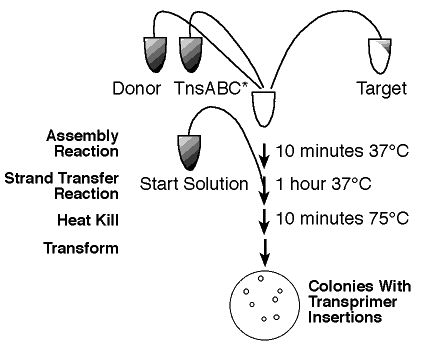
Step-by-Step
Simply isolate 'Your Favorite DNA' to be sequenced and follow the reaction
protocol provided by New England BioLabs Inc. (1) to create fragments ready
for sequencing. The plasmids lack the origin of replication (ori-), so
"only products (target DNA molecules containing Transprimer insertions)
can be recovered." (1) The company provides two transposons to be
inserted, one with kanamycin resistance and the other chloramphenicol resistance.
Therefore, you have the two controls to ensure that only plasmids with
transposon insertions are replicated.
Mixing TnsABC Transposase, an enzyme which inserts the transposons
into the target DNA, the target DNA, and the appropriate buffer is all
thatís necessary to create a Genomic Library using the provided
plasmid vectors. Once the vectors are transformed into competent
E. coli cells that are sensitive to the antibiotic, to which the chosen
transposon has a resistance, simply plate the cells with "drug combinations
appropriate to select for the target plasmid alone and for the target with
a transprimer insertion" (1). A plate should have around "100 colonies"
which is a sufficient number to complete the sequencing (1). Using
the provided primers (Primer N and Primer S) both strands of DNA can be
sequenced.
Figure 2. (1)
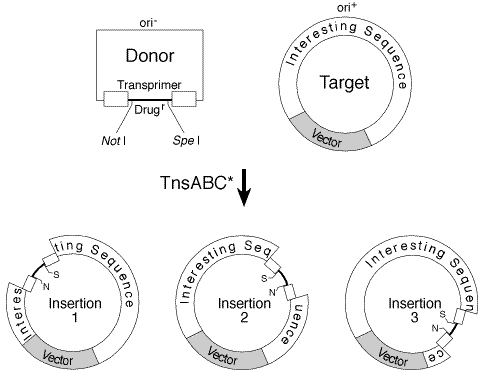
Controls
The Tn7 transposon carries a special immunity from Transposase.
The enzyme will not insert the vector into target DNA if it already contains
an insertion. Therfeore only one insertion of a tansposon will be
made per target DNA molecule.
Because of the ability to choose a drug resistance, only colonies
with insertions will be replicated.
Advantages
Fast. Instead of the old methods of preparing DNA for sequencing
like primer walking, where only short segments of DNA could be sequenced
at a time. The GPS-1 Genome Priming System allows an entire Genome
to be prepared for sequencing in under 90 minutes.
1) Easy. The protocol provided by BioLabs Inc. is easy to follow and
requires no special equipment that could not be found in your basic molecular
biology laboratory.
2) Controls. The convenient design of the plasmid a transposon
allows the researcher to selectively choose drug resistance and guard against
double insertions.
3) Efficient. 100-10000 "insertion products are recovered per
reaction."
References
1. "GPS-1 Genome Priming System." New England BioLabs Inc. Home Page.
11 Feb. 1999. 14 Feb. 1999. <http://www.neb.com/neb/tech/seq_and_label/7100.html
To return to your favorite web page click HERE

