PRO Bacterial Expression System
The PRO Bacterial Expression System is a method by which
one can tightly regulate expression of a protein, yet still induce a good
yield of that protein from a bacterial plasmid. This method is unique
because it used to be that when one wanted to tightly control the expression
of a gene there was always a decrease in the amount of protein for which
that gene coded. Thus, when one tightly regulated the gene one could
expect a low yield of its protein, while if one wanted a high yield of
it's protein one had to sacrifice control of that gene's expression, which
could potentially result in inclusion bodies within the cell ( inclusion
bodies are protein packets that accumulate when too much of a particular
protein has been made by the cell and the cell cannot use all the protein).
However, with the advent of the PRO Bacterial Expression System it is possible
to attain good yields with little protein leakage. The method is
also perfect for the expression of toxic proteins that have the potential
to damage the cell, because the PRO system prevents leakage of these proteins.
As an added bonus, this method even allows for the induction of two different
proteins at the same time.
The backbone(s) of the technique are the two plasmid
vectors, PROLar and PROTet. PROTet can induce production of the target
protein at a level that is 10% of total protein production. It is
the more inducible of the two vectors. However, PROLar is more tightly
controlled than PROTet, yet still maintains a respectable induction rate
of 2% of total protein made. Thus, PROLar is better suited for production
of toxic proteins, while PROTet is better suited for expressing a lot of
a non-toxic protein.
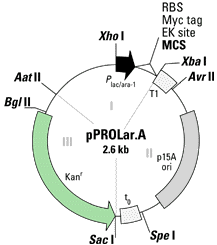
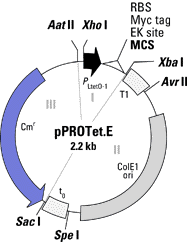
Each vector contains three components that
can be removed by restriction enzymes. Component 1 contains the promoter
region, the multiple cloning site, the ribosome binding site, and the Myc
tag for protein purification. Component 2 contains the origin of
replication and the transcriptional terminators, T1 and t0. Component
3 contains the antiobiotic resistance gene. The multiple cloning
site, the ribosome binding site, the Myc tag, and the transcriptional terminators
are conserved for both plasmids. However, the promoters, the ori,
and the antibiotic resistance gene are different for the two plasmids.
Finally, each plasmid vector comes in three different reading frames so
that the gene sequence being inserted is sure to be in the right reading
frame. The following are two cartoons of the plasmid vectors.
 (Images courtesy of CLONTECH
Labs, Inc.)
(Images courtesy of CLONTECH
Labs, Inc.)
In pPROTet the origin of replication is ColE1 and
the promoter is PLtetO-. ColE1 is a high-copy ori that has a 2,500
fold range of inducibility, meaning one can control protein induction over
a wide range by varying the concentration of promoter. The Pltet0-
promoter is a tetracycline-regulated promoter. This means it only
produces protein when it is "turned on" by anhydrotetracycline. The
promoter is further regulated by the Tet repressor, which allows for even
greater control of protein induction by turning the gene off (preventing
transcription) when it is present. Thus the highly inducible ColE1
ori and the tightly regulated promoter PLteto- together provide a plasmid
that can create many proteins, but only in the presence of anhydrotetracycline.
In pPROLar the origin of replication is p15A ori,
and the promoter is Plac/ara-1. The p15A ori is not as inducible
as the ColE1 ori in PPROTet, thus its low copy number decreases the chance
of protein leakage. The Plac/ara-1 promoter is based on the lac promoter
and is activated by the proteins Arabinose and IPTG. However, Plac/ara-1
is further controlled by a second lac operator sequence that is required
to allow RNA polymerase to transcribe the nucleotide sequence, thus providing
tighter control of protein induction. For this reason, the
pPROLar vector is best suited for the expression of toxic proteins. PPROLar
can repress the production of these toxic proteins until the cell has enough
materials to quickly produce them in sufficient amounts, then transcription
can be turned off so the toxic proteins don't further harm the cell.
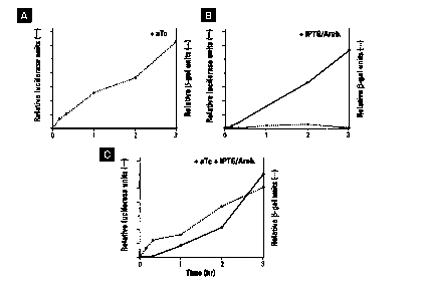
Since the the two plasmids use two different promoters
and two different origins of replication one can regulate protein induction
independently, allowing for induction of two proteins at the same time
in the same DNA strain. For example, if one uses only anhydrotetracycline
then only pPROTet is going to produce protein. However, if one uses
only IPTG/arabinose then the opposite is true, only pPROLar is going to
produce protein. Finally, if one uses both aTc and IPTG/Arabinose
then both proteins will be expressed. The following three figures
show protein that is expressed only by pPROTet, only by pPROLar, and both
pPROTet and pPROLar at the same time, respectively.
 (Images courtesy of CLONTECH
Labs, Inc.)
The Myc tag can be used to isolate your protein by using
a monoclonal antibody attached to a bead specific for that protein.
The protein is the only substance in solution that bonds to the beads,
so it can be separated from all other cellular machinery that is in the
solution. Then, one can break the hydrogen bonds holding the antibody
to the protein, and the protein can be run on a flourograh. This
technique is called immune precipitation.
(Images courtesy of CLONTECH
Labs, Inc.)
The Myc tag can be used to isolate your protein by using
a monoclonal antibody attached to a bead specific for that protein.
The protein is the only substance in solution that bonds to the beads,
so it can be separated from all other cellular machinery that is in the
solution. Then, one can break the hydrogen bonds holding the antibody
to the protein, and the protein can be run on a flourograh. This
technique is called immune precipitation.
Of course, the advances that this method offers are useless
if one doesn't know how to insert a nucleotide sequence into a plasmid, or even
why one would want to do this. The PRO Bacterial Expression System is
used when one wants to clone a certain nucleotide sequence and express the protein
for which this nucleotide sequence codes. For example, if one wanted to
clone the nucleotide sequence for the produtction of luciferase, then one would
find this gene sequence and use a restriction enzyme to cut both the bacterial
plasmid at the MCS and the nucleotide sequence out of a DNA strand. One
could use either pPROTet or pPROLar, it doesn't matter. Then, after making
sure the gene sequence being inserted is in the appropriate reading frame, the
plasmid is mixed with the DNA fragment of interest and the enzyme DNA ligase
joins the two strands together. Since there are many restriction sites
for a given restriction enzyme throughout an organism's genome and it is possible
for these other sites to bond with the plasmid or even to each other, many combinations
of plasmid and gene sequence are possible. To determine which plasmids
bonded to the gene of interest, we transform (term used to describe the ability
of bacteria to take up naked DNA) the plasmid into a bacterial cell, in this
case E. coli with an antibiotic resistance gene. Both pPROLar and
pPROTet have antibiotic resistance genes, so when placed on a growth medium
with an antibiotic, only these two plasmids with immunity to that antibiotic
will survive. Furthermore, one can analyze the restriction sites of the
plasmids that survived the antibiotic laden medium in order to confirm the orientation
of the insert. The Clontech procedure also recommends sequencing
to confirm the orientation of the insert. The final step is to induce
protein growth by adding the proper concentration of either anhydrotetracycline
for pPROTet or arabinose/IPTG for pPROLar, and purify your protein product by
using immuno precipitation or SDS-PAGE
and Western
blotting. To see a cartoon about cloning into a plasmid, go here.
The PRO Bacterial Expression system is an exciting
and innovative new technique that makes controlling protein induction much
easier.
Sources:
Campbell, N. A. 1996. Biology, Fourth Edition. Menlo
Park, CA: Benjamin/Cummings Publishing Company. p. 344-345.
"PRO Bacterial Expression System User Manual." 1998. CLONTECH
web resource-manual. <http://www.clontech.com/clontech/Manuals/PDF/PT3161-1.pdf>
Accessed Feb. 15, 1999.
"Cloning Genes." 1998. MIT-Web Resource. <http://esg-www.mit.edu:8001/bio/rdna/cloning.html>
Accessed Feb. 15, 1999.
Canfield, Elizabeth. "Sanger Method for DNA Sequencing."
1999. Lisa Canfield's webpage. <http://bio.davidson.edu/Biology/Courses/Bio111/seq.html>
Accessed Feb. 16, 1999.
White, Brian. "Southerns, Northerns, Westerns, and Cloning:
Molecular Searching Techniques." 1995. MIT web page. <http://esg-www.mit.edu:8001/bio/rdna/rdna.html#transfer>
Accessed Feb. 16, 1999.
"SDS-PAGE (Polyacrylamide Gel Electrophoresis)." 1998. Davidson
web page. <http://bio.davidson.edu/Biology/Courses/Molbio/SDSPAGE/SDSPAGE.html>
Accessed Feb. 15, 1999.
This webpage is for educational purposes only. To purchase this
product, click here.
To return to my homepage, click here.
email me: schutchins@davidson.edu



