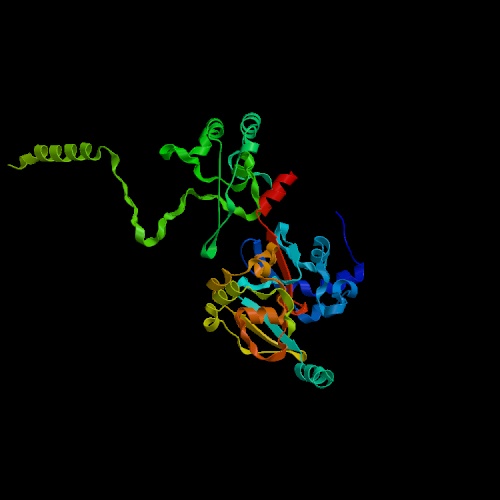
 Still image of the cyclin protein.
Permission requested from PDB:1QB3
Still image of the cyclin protein.
Permission requested from PDB:1QB3This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Gene(s)!
Annotated Gene: CCL1/YPR025C (YPD Protein Report)
Non-Annotated Gene: YHR078W (YPD Protein Report)
Annotated Gene: CCL1/YPR025C (YPD Protein Report)
Saccharomyces cerevisiae Chromosome XVI
SGD: 50006229 Genbank: CAA89279.1 MIPS (PIR): 539383 SWISS-PROT: P37366
Cyclin CCL1 serves several functions within the yeast as a component of TFIIK, the C-terminal domain (CTD) kinase subcomplex of TFIIH, which is required for transcription in vitro. The protein is missing, however, in another form of TFIIH which performs NER, nucleotide excision repair (Svejstrup, 1995). When introduced to RNA Polymerase II transcription and NER at the same time and mixed with the RAD26 gene, TFIIH chooses to be used in NER and thus inhibits transcription (You, Z., WJ Feaver et. al, 1998). CCL1 and KIN28 together make up TFIIK. KIN28, which is involved with the largest subunit of RNA Polymerase II, works physically and genetically with CCL1, and has been shown to be cyclin-dependent kinase. Without CCL1, some mutants were shown to be under-phosphorylated by simply KIN28, thus controlling mRNA transcription (Valay JG, MF Dubois, et al, 1996). This CTD kinase-cyclin pair stimulates the promoter clearance and transcriptional elongation, which leads to an accumulation of G1 cells and an increase in cell size (Ohkuni K and I. Yamashita, 2000).
 Still image of the cyclin protein.
Permission requested from PDB:1QB3
Still image of the cyclin protein.
Permission requested from PDB:1QB3
A deletion of this gene is lethal, but there are other mutations that exist which are viable. Two mutatants were created using PCR mutagenesis (ccl1-ts3 and ccl1-ts4) and were shown to exhibit heat sensitivity at 35 and 37 degrees, respectively. The ccl1-ts3 mutations proved to stop growth at 35 degrees, which indicates that CCL1 is required at various points during the cell cycle. At 37 degrees, the level of mRNAs decreases rapidly for the ccl1-ts4 mutation, but a subset of these mRNAs remains, which indicates that CCL1 may not be required for the transcription of all genes. Both the mutations and wild types showed a similar sensitivity to UV irradation (YPD Protein Report, Valay JG, MF Dubois, et al, 1996).
Non-Annotated Gene: YHR078W (YPD Protein Report)
S. cerevisiae Chromosome VIII
SGD: S0001120 Genbank: AAB68887.1 MIPS (PIR): S46809 SWISS-PROT: P38799
YHR078W is a hypothetical Open Reading Frame (ORF) whose biological function and cellular role are unknown, as well as the exact localization within the cell. Based on a BLASTp of the protein sequence, the nearest alignment comes from a hypothetical protein FLJ14972 encoded by a Homo sapiens gene, but there is only a 54% positives match, and the e-value is 2.7, which does not suggest a high likelyhood of homology. The YPD Protein Report offers a 56% relation to a predicted protein fragment encoded by a Kluyveromyces lactis ORF (GB:AJ229892), but this does not appear with a Blast search. These two specific yeast species show conserved gene order relationships as well as a unique homology between genes that is closer than between others species, which may become more significant as more discoveries are made (Ozier-Kalogeropoulos, O, A. Malpertuy et al., 1998).
According to a Kyte-Doolittle
analysis, there is a good chance that if this gene encodes for a protein, it
is transmembranic at several places: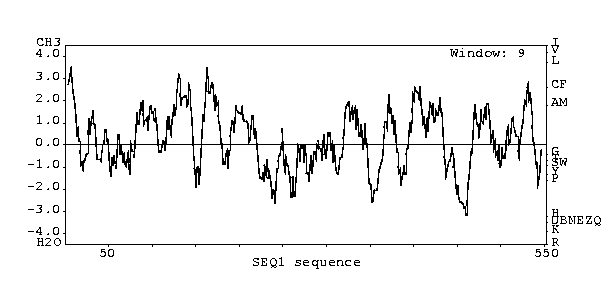
J. Kyte and R. F. Doolittle (1982) J. Mol. Biol. 157:105-132. Permission requested.
According to PREDATOR, another structure prediction method, this protein consists of alpha helicies (45.47%), random coils (41.12%), and extended strands (13.41%).
Based on its possible similarity to the Kluyveromyces lactis ORF, which according to a BLASTn search is a partial match with the Ovibos moschatus kappa-casein gene, although with an e-value of 0.22. The kappa-casein gene (in Homo sapiens) is a mammalian milk protein to aid in digestion (NCBI CD Browser), which could translate into a similar function for K. lactis or S. cerevisiae. This gene may not code for any kind of protein at all, but its homology with K. lactis suggests that it probably does, although we do not know anything about it yet.
References
(1) Svejstrup JQ, Wang Z., et al. 13 January, 1995. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell, Vol 80(1): 21-8. 29 September, 2001. <http://www3.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&uid=95112346&dopt=r>
(2) You, Zhaoyang, William J. Feaver, and Errol C. Friedberg. May 1998. Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inbibitors by Holo-TFIIH and requirement for RAD26. Molecular Cell Biology, Vol. 18(5):2668-76. 30 September, 2001. <http://www3.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&uid=98226539&dopt=r>
(3) Valay JG, MF Dubois et al. March 1996. Ccl1, a cyclin associated with protein kinase Kin28, controls the phosphorylation of RNA polymerase II largest subunit and mRNA transcription. C R Acad Sci III, Vol 319(3): 183-9. 30 September, 2001. <http://www3.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&uid=96340874&dopt=r>
(4) Ohkuni, K, and I. Yamashita. 30 June, 2000. A transcriptional autoregulatory loop for KIN28-CCL1 and SRB10-SRB11, each encoding RNA polymerase II CTD kinas-cyclin pair, stimulates the meiotic development of S. cerevisiae. Yeast, Vol 16(9): 829 - 46. 29 September, 2001. <http://www3.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&uid=10861906&dopt=r>
(5) Ozier-Kalogeropoulos, O., A. Malpertuy et al. 1 December, 1998. Random exploration of the Kluyveromyces lactis genome and comparison with that of Saccharomyces cerevisiae. Nucleic Acids Research, Vol 26(23): 5511 - 24. 30 September, 2001. <http://www.ncbi.nlm.nih.gov/entrez/utils/qmap.cgi?uid=99045620&form=6&db=m&Dopt=r>