Preface:
This web page lists sequence information for my favorite annotated
and nonannotated Saccharomyces cerevisae (yeast) gene. Each gene is
defined by its biological process, biological function and cellular components
as defined by Gene Ontology.
My favorite annotated yeast gene is KGD1, which codes for a subunit of the alpha ketogluterate dehydrogenase protein complex. This enzyme is responsible for the decarboxylation of alpha-ketogluterate in the tricarboxylic acid cycle (TCA cycle) and the reduction of NAD+. My favorite nonannotated yeast sequence is found in the same chromosomal region as KGD1 and is designated as YIL127C. Using databases and other web pages I will try and predict YIL127C’s function and possible roles in the life of yeast.
KGD1:
Biological Process:
The KGD1 gene encodes for 2-oxoglutarate dehydrogenase, a subunit in the alpha-ketogluterate
hehydrogenase complex, which is responsible for the decarboxylation of alpha-ketogluterate
in the TCA cycle. This enzyme plays an integral role in cellular respiration
and the production of ATP particularly since the enzymatic reaction is coupled
to the reduction of NAD+. The enzyme also plays an integral role in amino acid
metabolism.
Molecular Function:
The alpha-ketogluterate dehydrogenase complex is responsible for the catabolic
metabolism of alpha-ketogluterate to succinyl-CoA and CO2. The reaction requires
the cofactor thiamine pyrophosphate and the dehydrogenase complex consists of
three different subunits, 2-oxogluterate dehydrogenase (E1), dihydrolipoamide
succinyltransferase (E2) and lipoamide dehydrogenase (E3) encoded by KGD1, KGD2
and LPD1 respectfully. The reaction also requires the cofactor thiamine pyrophosphate.
Cellular Component:
2-oxoglutarate dehydrogenase and the alpha-ketogluterate deydrogenase complex
localizes in the mitochrondrial matrix. (SGD,
Swiss Prot, (Mockovciakova,
1993))
Physical Map for KGD1:

Figure 1: Physical map which spans 10 kb upstream and downstream of KGD1 on chromosome 9 (coordinates 112689 to 135733). KGD1 is also referred to as locus YIL125W.
KGD1
Coding Sequence
KGD1
Genomic Sequence
KGD1
Protein Translation
Mammalian
Homologs
Note: Thes results for mammalian homologs are not unexpected. The first three homologs with the highest high score and probability are for human alpha-ketogluterate. However, the next blast hits are for rat and mouse pyruvate dehyrogenase and as you scroll down the list you notice a slew of mouse immunoglobin proteins.
A PDB search for KGD1 yielded zero hits. However, when searching under 2-oxogluterate deydrogenase there were ten hits. The following chime image is for Lipoamide Dehydrogenase (E3) chain A and B (see figure 2).
Figure 2: PDB file for yeast Lipoamide Dehydrogenase (E3) chains A and B which
is part of the 2-oxogluterate dehydrogenase complex. The PDB database does not
currently have a structure for KGD1 (2-oxogluterate dehydrogenase) or the alpha-ketogluterate
dehydrogenase protein complex. The latter result is not surprising since the
complex consists of three subunits and is over at least 100 kilodaltons.
Additional Protein Information:
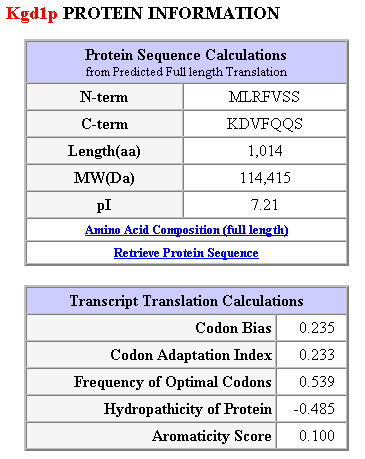
Table 1: Additional protein information and transcript translation calulations. This table was obtained from the following url: SGD Database kgdp1.
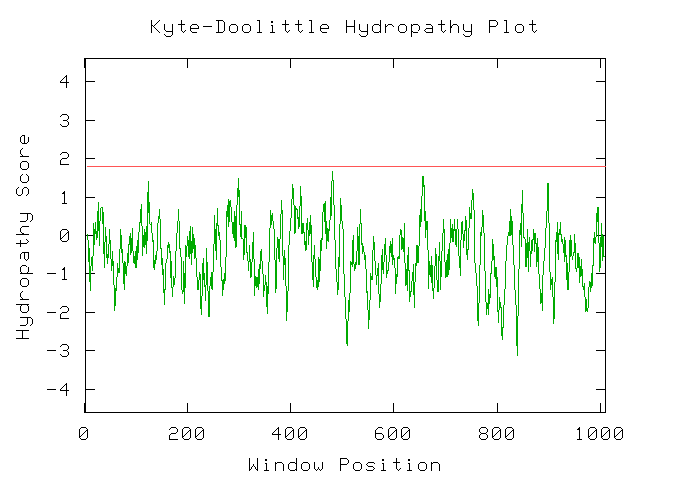
Figure 3: Kyte-Doolittle hydrophobicity plot for Kgd1. This indicates that Kgd1 does not have any transmembrane domains. I would expect these results since Kgd1 is a compent of the mitochondrial matrix.

Figure 4: Secondary structure predictions for KGD1 using PREDATOR. Each colored region denotes a different secondary structure: alpha helicies are colored blue (31.56%), extended strands are colored red (10.54%), and random coils are colored yellow (57.99%).

Figure 5: Conserved
domains for Kgd1. The first hit (E1_dehydrog) is for a family of proteins
with thiamine pyrophosphate cofactors (CD-Length = 301 residues, 96.0% aligned,
Score = 168 bits (426), Expect = 1e-42 ) .Other proteins in this family are
pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase and 2-oxoisovalerate dehydrogenase.
The second hit is for a transketolase, pyridine binding domain. Other proteins
in this family include transketolase enzymes, pyruvate dehydrogenases, and branched
chain alpha-keto acid decarboxylases. Now I can draw some conlcusions. Yeast
KGD1 is related to mamalian homologs by these cofactor domains. However, I still
can't explain why KGD1 has homology to mamalian immunoglobin proteins. One hypothesis
might be that immunoglobins and KGD1 are homologous in amino acid sequences
that are responsible for dimerization and trimerization. But let's return to
KGD1.....
According to SGD’s webpage, 2-oxogluterate dehydrogenase interacts with 13 different proteins (only some are listed here): KGD2, LEA1, UTP9, DBP8, LPD1, HPD1. For a complete list of proteins click here. Again, this result should not be unexpected. KGD1 is involved in a metabolic pathway - the TCA cycle- that involves numerous proteins.
Mutations and Phenotype:
So what happens to a yeast with a "bad" KGD1? In the yeast's case the systematic deletion of KGD1 results in a viable yeast (Giaever, 2002) while a null mutation results in a yeast that lacks alpha-ketogluterate deydrogenase, is “respitory deficient”, will not grow on glycerol and increases organic acid production when grown on glucose (Mockovciakova, 2002). Additionally, the activity of yeast 2-oxogluterate dehydrogenase can be reduced by two allelic mutations: ogd1 and kgd1 (Mockovciakova, 2002). Click here for reference url (look towards the bottom of the page).
What happens to a human with a "bad" KGD1? A search at OMIM yielded zero hits for human diseases associated with 2-oxogluterate dehydrogenase or KGD1. This may be explained by two possible results. Either the OMIM database is incomplete (no one has discovered a disease associated with KGD1 or the alpha-ketogluterate dehydrogenase) or any mutation in the protein that decreases the activity of the complex does not allow the fetus to come to term. As an interesting side not and addendum, seventy three different SNP's have been documented at the NCBI database for 2-oxogluterate dehydrogenase.
YIL127C:
This ORF is located on yeast chromosome IX between nucleotide 117644 and 117024.
SGD does not associate a molecular function or biological process with YIL127C.
A search of Pubmed and SGD literature databases yielded zero hits for YIL127C.
However, utilizing online resources and databases, I have retrieved the following
information concerning YIL127C:
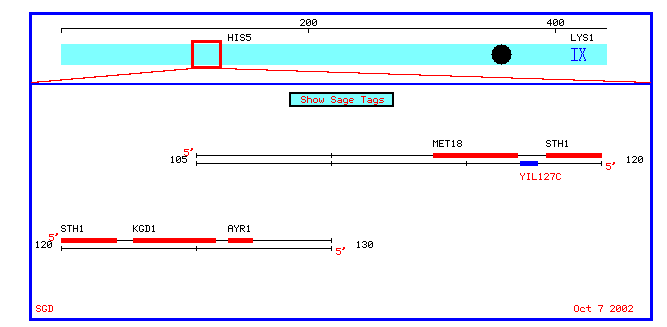
Figure 6: A chromosomal map of annotated and nonannotated genes in a 10 kb proximity to YIL127C on chromosome IX.
ATGTCATCTTCGCTGAGCCAAACATCTAAATACCAGGCCACCTCGGTTGTTAATGGACTG
CTATCAAACCTTTTGCCTGGCGTTCCCAAAATCAGAGCTAATAATGGCAAAACAAGTGTA
AACAATGGGTCAAAAGCTCAGTTAATCGACAGGAACTTGAAAAAGAGAGTACAGTTACAA
AACAGAGACGTTCACAAAATTAAAAAAAAATGCAAACTGGTCAAAAAAAAAAAAGTTAAG
AAACATAAATTAGATAAAGAGCAACTTGAACAACTGGCAAAACACCAGGTTTTGAAAAAA
CATCAACATGAGGGCACATTAACGGACCATGAGAGGAAGTATTTGAATAAGTTAATCAAA
AGGAATTCTCAAAATTTGAGGTCGTGGGATTTAGAAGAGGAAGTACGAGATGAGCTTGAA
GACATTCAGCAATCCATCTTGAAGGATACAGTTTCCACTGCGAACACAGACAGGAGTAAA
AGAAGAAGGTTCAAAAGAAAACAATTCAAGGAAGATATCAAAGAGAGCGATTTTGTTAAA
GATCATAGATATCCTGGTCTGACACCGGGTTTGGCACCTGTAGGACTAAGCGACGAGGAA
GACTCTAGTGAAGAAGATTAA
Figure 7: Here is the genomic sequence for YIL127C and the sequence is listed at the following url: http://genome-www4.stanford.edu/cgi-bin/SGD/getSeq?seq=YIL127C&flankl=0&flankr=0&map=a3map. Below is figure 8, which lists the corresponding amino acid translation otained from the SGD databases: http://genome-www4.stanford.edu/cgi-bin/SGD/getSeq?seq=YIL127C&flankl=0&flankr=0&map=p3map
MSSSLSQTSKYQATSVVNGLLSNLLPGVPKIRANNGKTSVNNGSKAQLIDRNLKKRVQLQ
NRDVHKIKKKCKLVKKKKVKKHKLDKEQLEQLAKHQVLKKHQHEGTLTDHERKYLNKLIK
RNSQNLRSWDLEEEVRDELEDIQQSILKDTVSTANTDRSKRRRFKRKQFKEDIKESDFVK
DHRYPGLTPGLAPVGLSDEEDSSEED
ORF: YIL127C has three open reading frames. One reading frame is 621 nucleotides long, one is 156 nucleotides long, and the other reading frame is 125 nucleotides long.
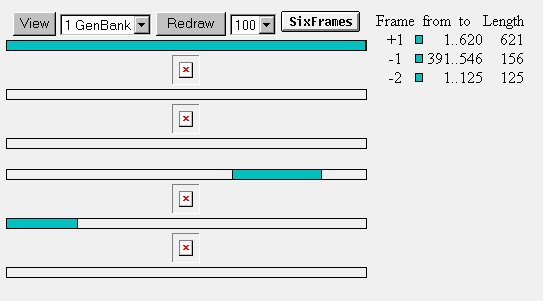
Figure 9: Open reading frames for YIL127C.
Figure 10: The blast search yielded four hits. The hit with the highest statistical similarity was a sequence on chromosome VII sequence with a high score of 175 and a sum probability of .92. The calculated identity and positives was 58%. This information is not as helpful for predicting YIL127C’s possible molecular function and biological process because the chromosome VII sequence is not annotated.
A yeast Blastp query resulted in few hits with low high scores and smallest sum probabilities. See Figure 11.
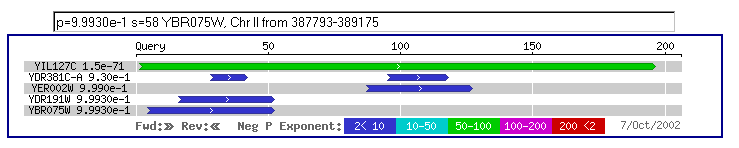
Figure 11: Keep in mind that the aforementioned blast search as well as this
one is only comparing yeast genes and yeast proteins. These searches are intended
to see how conserved YIL127C is in the genome.

Figure 12: A Fasta search of yeast DNA yielded several hits. The sequence with
the highest similarity had a bit score of 33.3 and an e value of .52. The identity
was 58.7% and 65% for ungapped. The similar sequence was ORF: Q0050/ AL1 and
which has a biological process of mRNA processing and is described as a “mobile
mitochrondrial group II intron of Cox1.” For more locus information click
here.
From this database we learn that YIL127C has homology to a Bovine ATPase inhibitor protein, which is only 84 amino acids long. YIL127C also has homology to a human V gamma T cell receptor protein, which is only 20 amino acids long.
Protein Predictions and Structural Information:
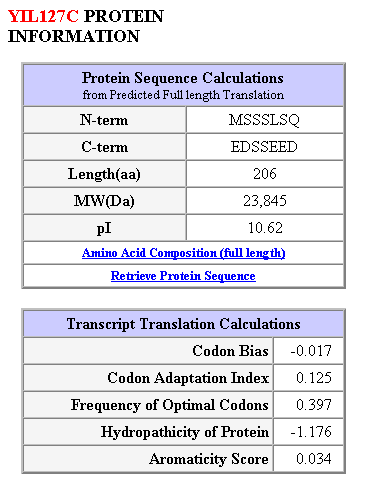
Figure 13: Protein structure and sequence calculations for the largest open reading frame (206 amino acids).
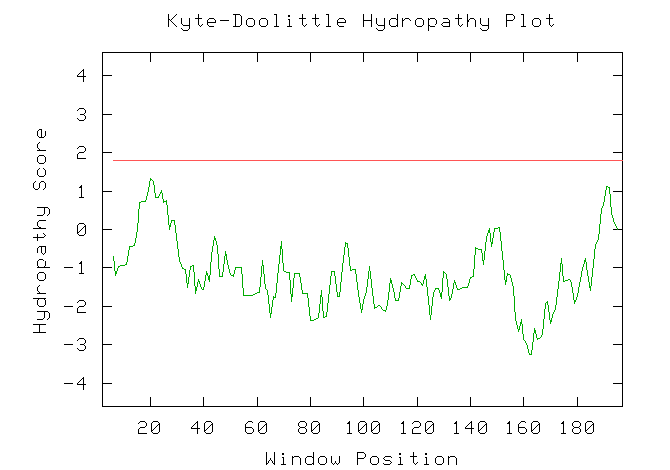
Figure 14: A Kyte-Doolittle hyrophobicity plot for YIL127C. The plot predicts that the theoretical protein for YIL127C does not contain any transmembrane domains.

Figure 15. Secondary structure predictions for YIL127C protein using PREDATOR.
The theoretical protein contains random coils (yellow, 54.37%), alpha helices
(blue, 42.72%) and extended strands (red, 2.91%).
The protein for YIL127C is small, only 29 kilo Daltons and does not contain any transmembrane domains. A search for conserved domains (CD) yielded zero hits. A blast comparison should determine the similarity between the bovine ATPase inhibitor and YIL127C. Right?
Figure 16: Blastp comparison for bovine ATPase homolog with YIL127C.
This is very perplexing, the SGD database lists the bovine receptor as a homolog to YIL12C but a blastp comparison finds no significant statistical similarity between the amino acid sequences. Nevertheless, this results is probably a consequence of more stringent parameters for the blastp algorithm.
Conclusions:
To recap: the SGD Fasta nt database found that YIL127C was similar to an intron
in Cox1; the SGD mammalian homolog database found similarity between a bovine
receptor, a Human V gamma T cell peptide and YIL127C. The large YIL127C ORF
is probably not an intron for Cox1 but it may serve as a functional mRNA that
is involved in mRNA processing. It is also possible that YIL127C encodes for
a bovine ATPase inhibitor since both proteins do not contain any transmembrane
domains according to a Kyte-Dolittle hyrophobicity plot (data not shown). The
Blastp results are troublesome because they suggest little to no similarity
between the bovine ATPase inhibitor and YIL127C. There were also no conserved
domain hits for YIL127C.
Predictions:
Biological Process: The data suggests YIL127C is probably an ATPase inhibitor.
Molecular function: The YIL127C probably forms a dimer with the cytosicl portion of yeast ATPase thereby inhibiting ATPase function.
Cellular Componets: YIL127C would probably be localized in the mitochrondrial matrix since the mitochondrial population density of ATPase is high relative to the rest of the cell.
References:
1. Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91 2002.
2. Mockovciakova, D et al. The ogd1 and kgd1 mutants lacking 2-oxoglutarate dehydrogenase activity in yeast are allelic and can be differentiated by the cloned amber suppressor. Current Genetics 24: 377-381 1993.
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28035
Send comments, questions, and suggestions to: alcubre@davidson.edu