This
web page was produced as an assignment for an undergraduate course at Davidson College.
Back
to Homepage.
My favorite yeast proteins
My search for information on Kex2 began at Mike Snyder’s TRIPLES
database. Unfortunately, this search
was fruitless. I then searched the PDB
in hopes of finding a CHIME image of the protein. This was also fruitless.
Stan Field’s yeast
two-hybrid data did not use Kex2 as bait, nor was it ever identified as
prey. My first positive result came
searching the DIP
database. The database returned an
interaction map shown below.
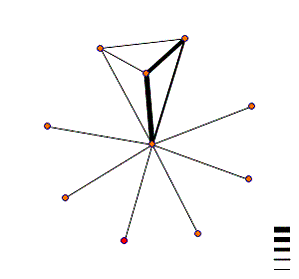
Kex2 is the red node, and is shown
to interact with one protein, identified as BFA1, though it should be noted
they use a thin line, indicating a low probability of actual interaction. A PubMed search returned abstracts
identifying this protein as being involved in cell cycle regulation(abstract here). This may suggest that Kex2 has some
additional role in cell cycle regulation.
The Curagen database indicates that Kex2 does not interact with any
other proteins(here). We know this is not true, but the database
is based on limited data. I then hoped
to identify Kex2 in a 2d gel database.
In the PROWL database, I found the MW of Kex2 to be 90000 and the pI to
be 4.8(here). Unfortunately, no spots were identified at
the corresponding points(gel shown here). There really isn’t much information
available about Kex2 in these databases.
To find out more about the Kex2 protein, I would start by performing a
yeast two-hybrid experiment using Kex2 as a bait protein to find out exactly
which proteins interact with it. We
would expect to find Kex2 interacting with the proteins it cleaves, but we
would also be looking to find proteins that may modify Kex2, or act as
coenzymes. I would then perform
experiments using the ICAT method, observing relative protein levels under
varying conditions. In particular, I
would want to verify the DIP data suggesting Kex2 has a role in the cell cycle
by observing relative Kex2 at different points in the cell cycle. I would also want to use the ICAT method to
determine if Kex2 is post-translationally modified in any way, such as
phosphorylation. Under the current
model of µ-factor
maturation, we would not expect to see any modification of Kex2. We would expect it to be active at all
times, its behavior being modified only by the presence or absence of its
substrate, the µ-factor.
SCYNL234W
is a somewhat more interesting story. Once
again, TRIPLES and PDB returned nothing.
The yeast two-hybrid system returned three hits however. All three are in the recent data link
from the main Y2H page. The first YNL234W
hit is as a prey ORF for CHK1 bait. A
PubMed search identified CHK1 as a kinase involved in the cell cycle arrest
response to DNA damage(here,
and here). The second hit is also as a prey ORF to the
bait protein CKB1. A PubMed identified
CKB1 as the regulatory subunit of caesin kinase 2(CKII), which has a few
different functions. Interestingly, one
of those functions is cell cycle control, specifically in exiting G1 and G2(here). The third hit is as a prey ORF for the bait
protein YPT31. PubMed revealed YPT31 as
a golgi protein involved in vesicular transport(here). It should be noted that two of the three
hits are involved in cell cycle regulation.
This may indicate a possible role of YNL234W in cell cycle signaling. Interestingly, the expression data did not
support this, though the limitations of those data have been noted. DIP identified YNL234W as interacting with
four different proteins, each involved in an intricate signaling pathway(click
on graph button in upper right of this link).
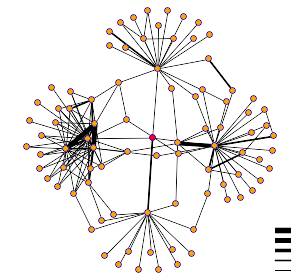
Three of the four identified
interactions(KC2B, YIV8, and IMB3) are shown with the thin bar, indicating a
low probability of interaction. One of
them is shown with the next thinnest bar.
This protein is CHK1, which Y2H also indicated. I then went to search Curagen, which indicated
that YNL234W did not interact with any other proteins(here). The PROWL database had not identified the MW
and pI of YNL234W, making it unidentifiable on their 2D gels. The cumulative data found in these sites
seem to indicate through a number of independent experiments that YNL234W may
play a role in cell cycle regulation.
This is quite interesting since none of our expression data or sequence
analysis posed this as a possibility, though they do not rule it out. One reason the expression data may not
suggest this is that expression levels may not be the primary activity of the protein. The proteins it appears to interact with are
kinases, leading us to conclude that the primary regulator of this protein may
be its phosphorylation state.
To
find out more, I would start by performing Y2H experiments with YNL234W as the
bait protein, in hopes of confirming previous data. I would then perform ICAT experiments examining protein levels,
and phosphorylation states at different points in the cell cycle, and also in
response to radiation damage to DNA. If
YNL234W is indeed involved in the cell cycle, as it appears to be, the most
critical data will probably be its phosphorylation state under these conditions,
given that the proteins with which it appears to interact are kinases.
Send comments, questions and suggestions to edhaas@davidson.edu
Many thanks to Dr. A. Malcolm Campbell for his guidance in this endeavor as well as others.