This web page was created as an assignment for an undergraduate course at Davidson College.
The Story of a Yeast Receptor Gene and an Unknown Neighbor
PART 3: YEAST PROTEINS
Note: This is the third part of a three-part exploration. To see information regarding these two genes, please go here.
Annotated Gene: STE2 (YFL026W)
As a quick summary, the main purpose of this page is to explore the protein data regarding two yeast genes: STE2 and a possible gene known as BUD27.
Below is the gene ontology for STE2 (Cherry et al., 1997):
Molecular Function: mating-type alpha-factor pheromone receptor (Burkholder AC and Hartwell LH, 1985)
Biological Process: pheromone response
Cellular Component: integral plasma membrane protein
I obtained the amino acid sequence of STE2 from the Saccharomyces cerevisiae Genome Database.
MSDAAPSLSNLFYDPTYNPGQSTINYTSIYGNGSTITFDELQGLVNSTVTQAIMFGVRCG AAALTLIVMWMTSRSRKTPIFIINQVSLFLIILHSALYFKYLLSNYSSVTYALTGFPQFI SRGDVHVYGATNIIQVLLVASIETSLVFQIKVIFTGDNFKRIGLMLTSISFTLGIATVTM YFVSAVKGMIVTYNDVSATQDKYFNASTILLASSINFMSFVLVVKLILAIRSRRFLGLKQ FDSFHILLIMSCQSLLVPSIIFILAYSLEPNQGTDVLTTVATLLAVLSLPLSSMWATAAN NASKTNTITSDFTTSTDRFYPGTLSSFQTDSINNDAKSSLRSRLYDLYPRRKETTSDKHS ERTFVSETADDIEKNQFYQLPTPTSSKNTRIGPFADASYKEGEVEPVDMYTPDTAADEEA RKFWTEDNNNL
To begin this protein exploration, I first went to the Database of Interacting Proteins (DIP). When STE2 is a node, I got the protein interaction graph shown below:

Figure 1: Protein interaction map for STE2 from DIP (DIP 2003). The red dot in the middle of the graph is STE2 and it connects through the lines to other proteins that it (may) interact with (orange dots).
The three proteins shown to interact with STE2 are listed below. The protein that appears to be in the middle of the much larger cluster is YSC84.
YSC84 is involved in actin filament organization, but this is an ORF of which very little is known, except its strong interactions with the other proteins in its cluster. STE2's interaction with YSC84 may spawn from the eventual morphogenesis of non-mating yeast cell to one that is ready to mate, taking on the shape of a shmoo (go here to see a picture of a shmoo). Actin reorganization could be needed for this to occur. YSC84 then may play a role in the yeast mating signal cascade.
YGR141w is an ORF that is possibly involved in protein-vacuolar targeting, but research is still being done on this. Little is known about this ORF.
SSP2 is a protein involved in spore wall assembly. It is located in the spore wall and may interact with STE2 since both are in the outermost membrane/wall of the cell.
In order to view the information pertaining to these proteins and to the interactions of STE2, you can go here. This webpage presents all the information about the proteins that interact with STE2. According to it, these interactions were detected using the Yeast Two-Hybrid system.
The Yeast PathCalling information page documented no protein interactions with STE2, which differs from the data obtained from SGD. If anything, one would think that STE2p interacts with other proteins (or at least one other protein) in the mating factor signal transduction pathway. However, seen here and in the protein interaction map above, no interactions pertaining to this pathway are noted.
The figure from the "Benno paper" did not document STE2 at all (Schwikowski, et al., 2000). Click here to see this figure as a PDF file.
No structural information could be obtained from the Protein Data Bank (PDB) either on this protein since it is an integral membrane protein.
Non-Annotated Gene: BUD27 (YFL023W)
BUD27 is not annotated, so the gene ontology information is not available. Exploring possibilities for the gene ontology is the primary goal of this webpage.
The amino acid sequence was found at SGD:
MDLLAASVESTLKNLQDKRNFLSEQREHYIDIRSRLVRFINDNDDGEEEGEGQGMVFGDI IISTSKIYLSLGYEYYVEKTKEEAITFVDDKLKLMEDAIEQFNLKIEEAKKTLDNLNHME DGNGIEEDEANNDEDFLPSMEIREELDDEGNVISSSVTPTTKQPSQSNSKKEQTPAVGPK EKGLAKEKKSKSFEENLKGKLLKRNDEVKKKVQPSKVDTENVYTFADLVQQMDQQDELED GYIETDEINYDYDAFENSNFKVNDNYEEDDEDEDEEEYLNHSIIPGFEAQSSFLQQIQRL RAQKQSQDHEREEGDVNKSLKPILKKSSFAENSDKKQKKKQVGFASSLEIHEVENLKEEN KRQMQSFAVPMYETQESTGIANKMTSDEFDGDLFAKMLGVQEADEVHEKYKEELINQERL EGEASRSNRRTRVSRFRKDRASKKENTLSTFKQETTRSVENEVVEKEPVVGDIIEKEPVV GDVIEKEPVVGDVIEKEPAVTDIVEREPAVNDIVERKPVVGDIIEKEPTINDIVEKEPEI NSKSEFETPFKKKKLKSLQKPRSSKSMKKKFDPKILENISDDDYDDDDDGNKKLLSNKSK NNTDEQDKFPSKIQEVSRSMAKTGATVGSEPVRITNVDYHALGGNLDDMVKAYSLGLYDD DLEEDPGTIVEKLEDFKEYNKQVELLRDEIRDFQLENKPVTMEEEENDGNVMNDIIEHEF PESYTNDEDEVALHPGRLQEEVAIEYRRLKEATASKWQSSSPAAHTEGELEPIDKFGNPV KTSRFRSQRLHMDSKP
When I viewed YFL023W in DIP, I obtained the following graph. It has five protein interactions and each of those proteins is documented below the image.

Figure 2: YFL023W protein interaction map from DIP. The red dot in the middle is YFL023W. It interacts with five other proteins (orange dots).
RPB5 is a DNA-directed RNA polymerase I, II, and III. It is involved in the RNA polymerase complex.
NFS1 is involved in iron homeostasis and it is located in the mitochondrion. This seems to be an unreasonable interaction since NFS1 is located in the mitochondrion and thus far, I have seen no other references localizing YFL023W to the mitochondrion. Its other protein interactions seem to be with proteins in the cytosol or nucleus.
YLR243w is an unknown ORF, but it is crucial for cell viability because when it is deleted, the cells are inviable.
YKE2 is a protein located in the cytoplasm that is involved in yeast tubulin folding and binding. (See below for a discussion of its implications with YFL023w).
CID1 is a busy protein involved in glycogen metabolism, meiosis, and mitotic spindle checkpoints.
The Saccharomyces cerevisiae Genome Database also provides a list of these interactions and documents what each protein does and its gene ontology information. To see this page, click here.
Below is information obtained from the Yeast PathCalling Home Page.
Yeast PathCalling Home Page (CuraGen & University of Washington)
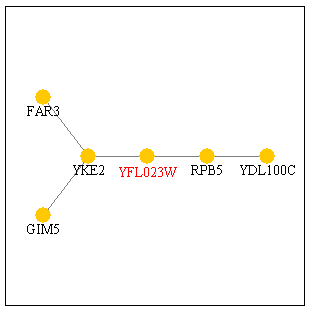
Figure 3: Protein interaction map for YFL023W from the Yeast Pathcalling Home Page. YFL023W is shown in red.
The two genes immediately connected toYFL023W are RPB5 and YKE2. YKE2 is located in the cytoplasm and is involved with tubulin and protein folding. RPB5 is part of the DNA-directed RNA polymerase complex and is involved in transcription, as noted above. If these two gene products interact with BUD27, then RPB5 could be the transcription factor that helps transcribe BUD27 and YKE2 could be a gene product that BUD27 acts on. If BUD27 is involves with budding, then it would make sense for it to act on or interact with a gene that is involved with tubulin and protein folding. Conversely, the protein encoded by BUD27 could be folded by YKE2, suggesting its possible tubulin properties? A closer look reveals that GIM5 is also a protein involved in protein folding. However, FAR3 is a protein needed to arrest the cells in G1 of the cell cycle in response to the mating pheromone. By looking at the microarray data above, I had ruled out BUD27's role in pheromone response or in mating. Perhaps, however, the interaction between BUD27 and FAR3 is a "negative" interaction. Maybe BUD27 is responsible for turning FAR3 "off." If BUD27 is a transcriptional regulator or transcript processor, then this suggestion would work.
The figure from the "Benno paper" did not document YFL023W at all (Schwikowski, et al., 2000). Click here to see this figure as a PDF file.
I explored YFL023w using Function Junction. It has no significant homologies to worm proteins and no significant results were obtained from the Yeast Triples Database.
Furthermore, I tried to do a structural analysis using NCBI's Structure Query and was unable to obtain a result in its structure database or in PDB.
Conclusion
I found out very little more when I used these databases. In lieu of this data and the data gathered on the yeast expression and yeast genomic pages, I have concluded that YFL023W is a transciptional regulator or processor that interacts with genes and/or proteins involved in the budding pathway. It could also act to turn off genes/proteins that may be involved in the mating pathway so the cell does not prepare itself for mating, but redirects its energy to the budding pathway.
How To Find Out More
In order to make a more conclusive interpretation, I have designed a couple of experiments. First, YFL023W is shown to interact with two separate proteins, RPB5 and YKE2, that are located in two different parts of the cell: the nucleus and the cytoplasm, respectively. There is no data that discusses the localization of YFL023W. It could be expressed in the nucleus or the cytoplasm or both, possibly at different times. In order to explore this, I would want to do a localization procedure to see where YFL023W can be found. Immunofluorescence is more of a molecular technique than a genomic or proteomic technique, but sometimes it's good to go back to basics when exploring the localization and/or expression of a single protein or gene. In this case, you can make a fluorescent probe using the known amino acid sequence of YFL023W (see above) and then you apply this probe to the cells in question. When the cells are under the correct light source, you can see where the probe bound. This should be the location of the protein. A proper control would need to be performed by showing that the probe does indeed bind to YFL023W and that it does not bind to any other protein (apply the probe to YFL023W deletants). There should not be any fluorescence. By performing this technique on wildtype cells in culture, I will be able to see where YFL023W is localized. I could also perform this technique during any number of other times in the cell cycle or under any type of conditions to see if the protein is localized in different cellular compartments at different times.
Another experiment could provide answers concerning the interactions of YFL023W if it was a transciptional regulator. I had concluded that YFL023W might be a regulator or processor that interacts with genes and/or proteins involved in the budding pathway. In the absence of mating factor, cells will bud in order to reproduce. I could isolate wildtype cells and perform a microarray analysis of the genes in this cell. I could then do the same for cells where YFL023W is deleted. This could distinguish candidate genes that might be regulated downstream from YFL023W or that might be regulated by YFL023W. If the genes are regulated transcriptionally by YFL023W, then when this gene is deleted, the expression profiles of these genes will be different compared to the expression profiles when YFL023W was present. Another possibility to look at is the expression of YFL023W expression and the expression of any genes that seem associated with it when wildtype cells are actually in the midst of budding. I could compare these expression profiles with the profiles of these same genes when mating factor is present to see if there is any difference in expression. Again, these microarray experiments would help provide a general overview, where molecular techniques would aid in narrowing down the search and providing more detailed information concerning specific genes in question.
References
Burkholder AC and Hartwell LH. 1985. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res 13(23):8463-75.
Campbell AM and Heyer LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. (The figure from the Benno paper was obtained through the webpage affiliated with this book).
Cherry, J. M., Ball, C., Dolinski, K., Dwight, S., Harris, M., Matese, J. C., Sherlock, G., Binkley, G., Jin, H., Weng, S., and Botstein, D. 1997-2000. "Saccharomyces Genome Database" http://genome-www.stanford.edu/Saccharomyces/ Accessed: 04 October 2002. *Function Junction and Expression Connection are interactive databases associated with SGD.
Chu S, et al. 1998. The transcriptional program of sporulation in budding yeast. Science 282(5389):699-705. (abstract)
Database of Interacting Proteins. http://dip.doe-mbi.ucla.edu/dip/Main.cgi
DeRisi JL, et al. 1997. Exploring the metabolic and genetic
control of gene expression on a genomic scale. Science 278(5338):680-6.
(abstract)
Schwikowski, B, Uetz P, and Fields S. 2000. A network of protein-protein interactions in yeast. Nat Biotechnol 18(12):1257-61. (abstract) (PDF file)
Sprague GF.
1991. Signal transduction
in yeast mating: receptors, transcription factors, and the kinase connection.
Trends Genetics 7(11-12):393-8.
Return Home
Return to the Genomics, Proteomics, and Bioinformatics Homepage
© Copyright 2002 Department of Biology, Davidson College, Davidson NC 28035
Send comments, questions, and suggestions to vistatler@davidson.edu
