"This web page was produced as an assignment for an undergraduate course at Davidson College"
MIF2 and YKL091C
MICROARRAY ANALYSIS
![]()
"This web page was produced as an assignment for an undergraduate course at Davidson College"
MIF2 and YKL091C
MICROARRAY ANALYSIS
![]()
Microarrays provide biologists with a powerful new tool for analyzing the gene expression of every gene in a given genome concurrently. By analyzing the reaction of the genome through regulation of gene expression under varying conditions, biologists can gain greater insight into the function and relationship of both known and unknown genes.
As discussed in the previous website assignment, the MIF2 gene encodes for a protein essential to the mitotic process in Saccharomyces cerevisiae. MIF2 encodes for a centromeric protein involved in mitotic spindle structural assembly. The MIF2 protein is responsible for maintaining the structural integrity of the spindle prior to anaphase spindle elongation. The information used to further analyze MIF2 using microarrays was supplied by the Expression Connection website.
Microarray Gene Induction/Repression scale
![]()
Expression of MIF2 in response to alpha-factor
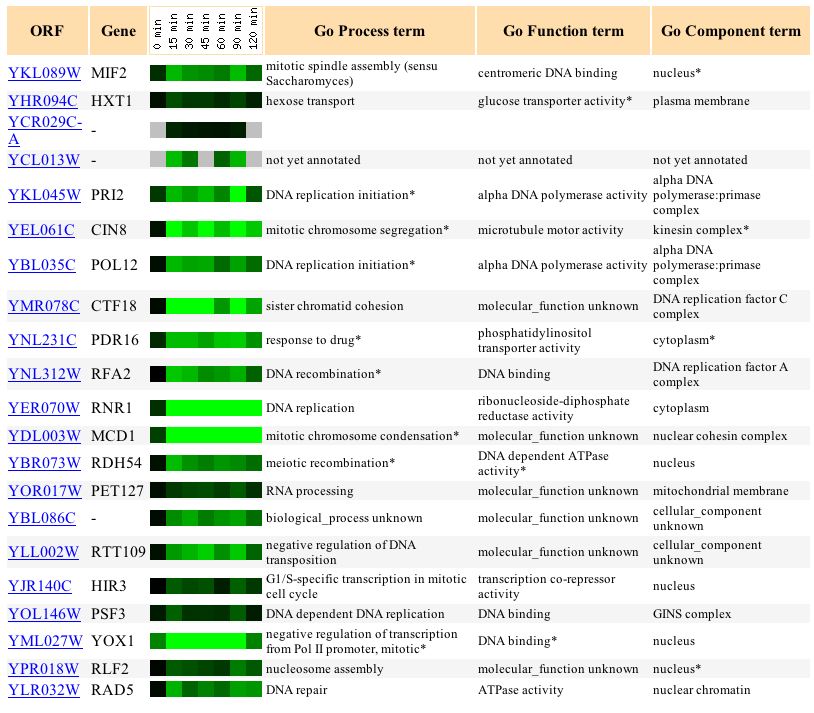
Figure 1. Screen capture of results from alpha-factor experiment. (Expression Connection)
The alpha-factor experiment reveals that MIF2 is decreasingly repressed as it is exposed to a constant amount of alpha-factor over time, with the exception of the 90min time interval in which the gene experiences a sudden increase in repression. It is interesting to note that the gene HXT1, whose expression pattern is most similar, functions in glucose transportation. This pairing demonstrates that there can be exceptions to the "guilt by association" rule. Because two genes are regulated similarly that does not necessarily mean that they have similar function. A possible reason for the pairing might be that MIF2 and HXT1 share a common promoter. On the other hand, the two genes PRI2 and CIN8, which also share a significant degree of expression with MIF2, appear to function in the mitotic process as well. PRI2 functions in the initiation of DNA replication and CIN8 functions in mitotic chromosome segregation. Alpha factor functions to halt mitosis in preparation for reproduction; therefore, it makes sense that a gene that encodes for a protein involved in the mitotic process is repressed.
Expression of MIF2 during diauxic shift
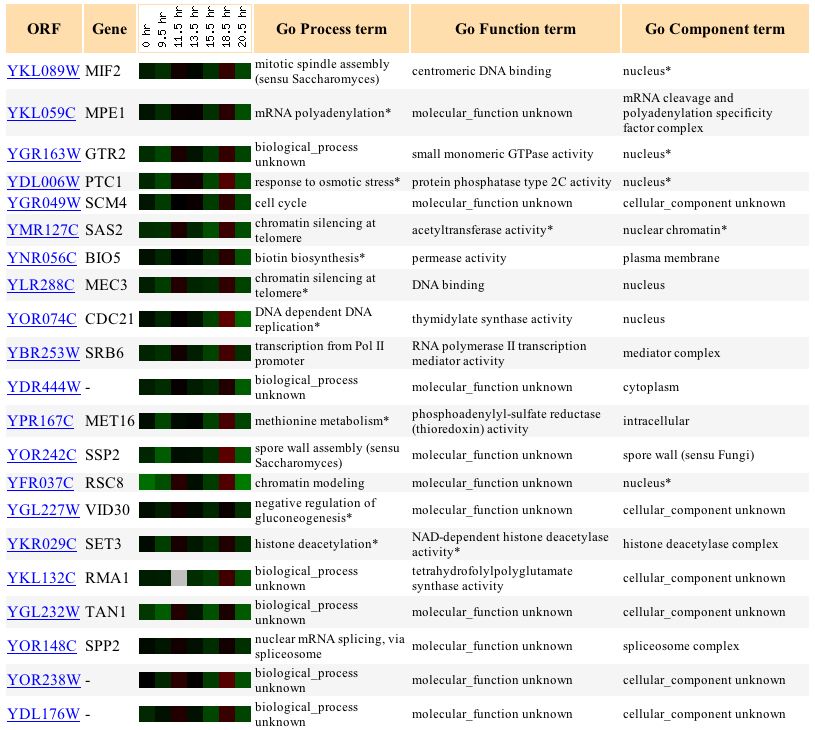
Figure 2. Screen capture of results from diauxic shift experiment (Expression Connection)
The expression pattern of MIF2 during the diauxic shift is highly varable. It experiences a low level of repression, with the exception of 11.5 hr. and 18.5 hr. when it experiences a low level of induction. The diauxic shift is the shift from anaerobic to aerobic respiration. The reason for the variability in expression is most likely due to the yeast's attempts to acclimate to the new system of respiration punctuated by attempts at undergoing mitosis under the new respiration system.
Expression of MIF2 during the cell cycle
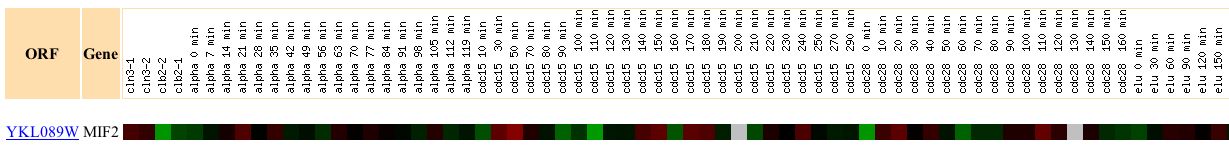
Figure 3. Screen capture of results of cell cycle experiment
(Expression Connection: Complete Figure)
In response to the cell cycle, the MIF2 gene undergoes a cycle of induction and repression. As stated previously, the MIF2 encoded protein is essential to the maintainence of the structural integrity of the spindle during mitosis. MIF2 expression is expected to be variable because the expression of the gene is dependent upon the stage in the mitotic cycle, and whether or not the spindle structure protein is required during a given step in that cycle.
Expression of MIF2 in response to environmental changes
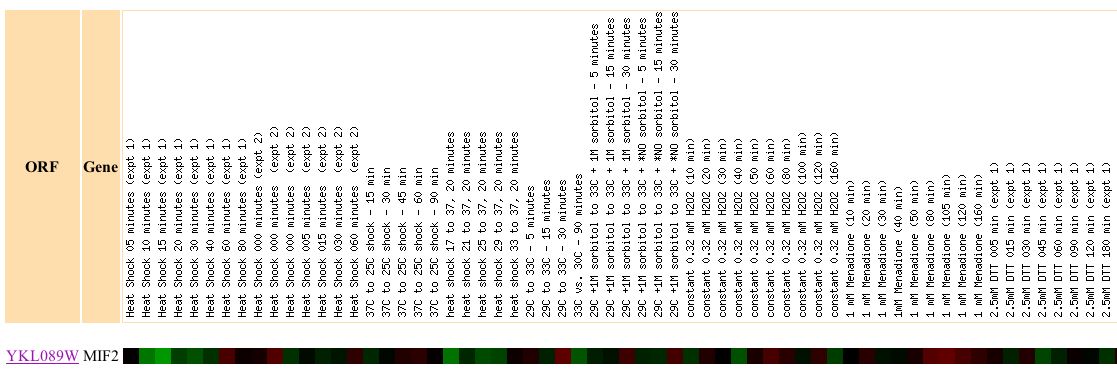
Figure 4. Screen capture of results of environmental experiments (Expression Connection: Complete Figure)
This figure depicts the expression of MIF2 depending during a number of changes to its environment, such as exposure to heat, hydrogen peroxide, menadione, DDT, and nitrogen depletion. Since the protein encoded by MIF2 functions in mitosis. We would expect the MIF2 gene to be repressed at the initiation of the shock, and induced once the yeast has had sufficient time to adapt to its new environment. We hypothesize that the the genome must respond to the shock and so represses gene expression involved in cellular replication in order to focus its energy on surviving the initial shock.For the most part this figure corroborates this hypothesis, we typically see repression of MIF2 during the initial stage of the shock and induction after a period of continuous exposure. Heat shock expt. 1 and 2 exhibit this trend in MIF2 gene regulation. There are also exceptions to this trend, however, conditions in which the mitotic pathway of which MIF2 is a part are directly affected by the stress. As appears to be the case in 1.5 mM diamide exposure and in nitrogen depletion. MIF2 gene expression is repressed throughout the course of the diamide shock and induced during the nitrogen depletion shock.
Expression of MIF2 in response to DNA-damaging agents

Figure 5. Screen capture of results of DNA-damaging experiments (Expression Connection)
This figure depicts the MIF2 expression response to DNA-damaging agents. MIF2 gene expression was induced the greatest at wt+gamma irradiation(10 min) and (60min). The greatest repression occured at wt+heat (20min), but then repression decreased as the genome adapted to the heat exposure as would be expected. At first glance, I was surprised by the results of these experiments. The genome responded to irradiation and other DNA-damaging agents by inducing a gene involved in mitosis. Why would the genome respond by undergoing mitosis with damaged DNA? One possibility for the induction of genes involved in the mitotic process is that the genome is trying to induce mitosis before the DNA becomes too damaged to do so.
________________________________________________
As discussed in the previous web assignment, the YKL091C gene was selected because it is an unannotated gene that lies in close proximity to the annotated gene MIF2. The research from the previous assignment did not yield a definitive answer as to the possible function of the YKL091C. However, there is a possibility that the protein encoded by YKL091C is a PITP, phosphatidylinositol transfer-like protein, involved in modulation of signal transduction and in transport across the membrane.
Microarray Gene Induction/Repression scale
![]()
Expression of YKL091C in response to alpha-factor:
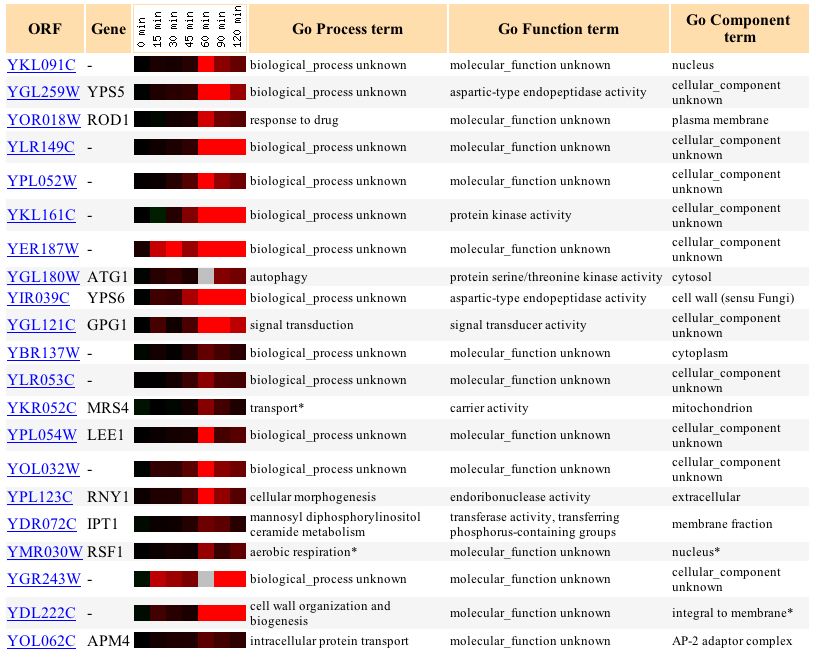
Figure 6. Screen capture of results of alpha-factor experiment
The YKL091C gene expression pattern consists of initial low-level induction, a sudden large increase in induction, which is then followed by a gradual decraease in induction. Most of the genes found to share similiarity in gene expression are predominantly of unknown function, with the exception of several less similar genes involved in transport and respiration. The fact that several related genes are involved in aerobic respiration (RSF1), signal transduction (GPG1), and transport (MRS4) support the conjecture that the protein encoded by YKL091C is a PITP, which is involved in transport (membrane trafficking) and transduction (signal transduction modulator). The information provided in this figure corroborates the hypothesis that YKL091C encodes for a PITP.
Expression of YKL091C during diauxic shift

Figure 7. Screen capture of results from diauxic shift experiment (Expression Connection)
This figure exhibits the dramatic change in YKL091C gene expression in response to the shift from anaerobic respiration to aerobic respiration. Initially, YKL091C is repressed under anaerobic respiration, but highly induced under aerobic respiration. Again, it is apparent that genes involved in aerobic respiration (QCR8), transport (EMP46), and signal transduction (MDG1) share similar expression patterns with YKL091C. Perhaps the YKL091C protein signals a pathway related to aerobic respiration or is reponsible for transporting the starting materials or products of aerobic respiration. YKL091C is an aerobic gene.
Expression of YKL091C in response to glucose limitation
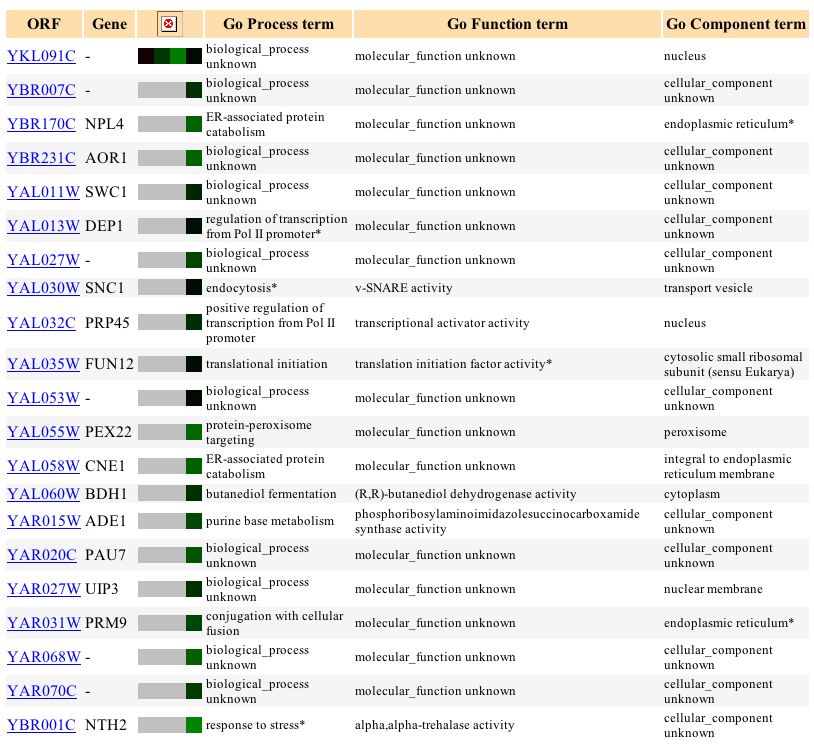
Figure 8. Screen capture of results of glucose limitation experiment (Expression Connection)
When yeast senses a depleting source of glucose it shifts to anaerobic respiration so that it can metabolize the glucose and store it faster before its neighbors have the chance to use it. As a result, aerobic genes such as YKL091C are consequently repressed in a glucose limited environment conducive to anaerobic respiration. This figure demonstrates that this is in fact the case with YKL091C, which experiences increasing repression under the condition of decreasing glucose availability. The final time interval indicates however that YKL091C is neither induced or repressed, which is most likely due to the absence of glucose completely and the impending switch from fermentation to ethanol metabolism.
Function Hypothesis
The expression patterns of the unannotated gene under the experimental conditions and functions of genes determined to share these patterns support my prior hypothesis that YKL091C encodes for a PITP. The genes with similar expression patterns were primarily of three functional types: signal transduction genes, membrane transport genes, and genes involved in aerobic respiration. In light of the microarray data, I hypothesize that the YKL091C is an aerobic gene that does in fact encode for a PITP, which functions in signal transduction and membrane trafficking.
Links:
References:
Expression Connection Search. Saccharomyces Genome Database. <http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?query=MIF2>
Expression Connection Search. Saccharomyces Genome Database. <http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?query=YKL091C>
"MIF2/YKL089W." Saccharomyces Genome Database. Last
Update: 2003. <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?sgdid=S0001572>
"YKL091C." Saccharomyces Genome Database. Last Update: 2003. <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=YKL091C>
© Copyright 2003 Department of Biology, Davidson College, Davidson,
NC 28035
Send comments, questions, and suggestions to: dacheuy@davidson.edu