This webpage was produced as an assignment for an undergraduate course at Davidson College
My Favorite Yeast Expression
YGR058W
Background: After determining YGR058W to be located on chromosome 7 I attempted to determine its structure, location, and function. The dearth of information available on this gene made determining these things difficult. Yet from my research into the location and function of YGR058W I found that the gene has some relationship to a gene called Saccharomyces cerevisiae sporulation specific septin (SPR3). This gene belongs to a family of bud neck microfilaments found in yeast. Thus I hypothesized that this gene is somehow related to the process of sporulation. Yet I needed more information to gain further insight into the gene's ontology.
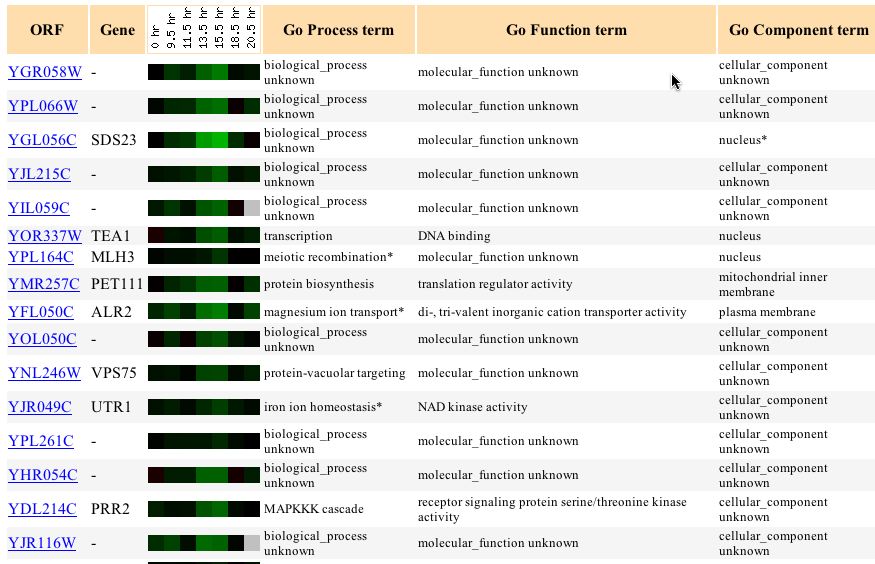
Using the Gene Expression databases I attempted to see under which experimental conditions YGR058W was induced or repressed. Then using that info I attempted to see if there were any genes with similar gene expression profiles. This method would allow me to associate this gene with other gene that have similar expression profiles--a method called "Guilt by Association". Unfortunately the results were not as clear as I hoped. The glucose limiting condition showed little difference in induction or repression and showed no similarity to other genes. The diauxic shift experiment showed an increasing level of YGR058W repression starting at 9.5 hours and continuing to 13.5 hours. This expression profile matched several other genes but, unfortuantely these genes were also undefined.

Figure 1 - Gene expression similarity to YGR058W when exposed to diauxic shift
The expression of YGR058W in response to alpha-factor over time and with various concentrations showed no change in expression profile. Thus, like with glucose limiting conditions, the cell does not change transcription levels of YGR058W with alpha factor induction of the cell. An interesting finding came from the expression profile with varying zinc levels. With varying levels of zinc the yeast genome induces greater transcription of YGR058W. More interesting is that genes with somewhat similar expression profiles have biological function related to cell proliferation, cell organization, and stress related ceulluar response (Figure 2).
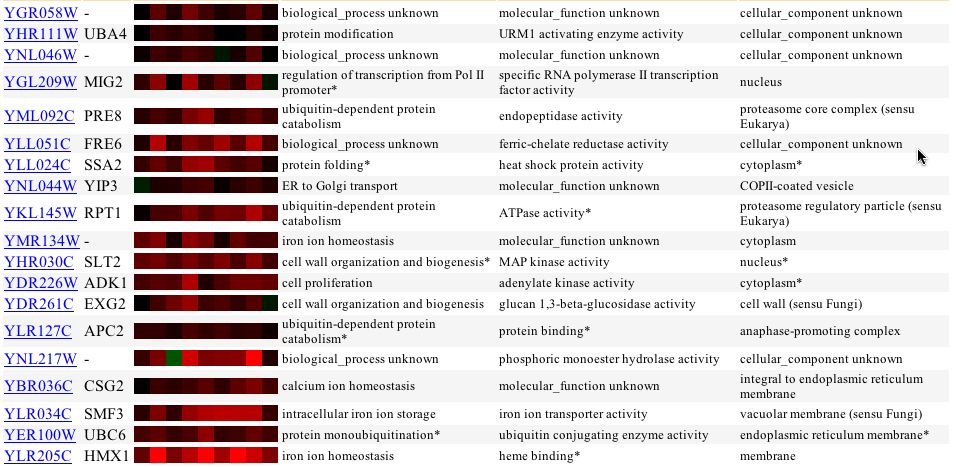
Figure 2 - Expression of YGR058W in yeast exposed to varying levels of zinc (First 4 are wildtype yeast with increasing zinc concentration. 5-7 are zap1 mutants)
The findings of the "expression regulated by PHO pathway" microarray data hinted at a similar function for YGR058W. This experiment showed what yeast genes are expressed under low phosphate concentration. Phosphate is an essential nutrient for various parts of life from nucleic acid formation to metabolic pathways (Phosphate Metabolism in Yeast). It is conceivable that low phosphate concentrations would induce genes involved in areas such as cell proliferation, cell organization, stress genes, and sporulation. Several genes had similar expression profiles as YGR058W. What caught my eye was YHB1 and ADK1 which are involed in response to stress and proliferation, respectively. While these two genes were not the closest in their expression similarity, they were pretty close and their involvement in stress and proliferation connected with the findings on the zinc. This gene seems more and more likely to be involved with something related to how the cell deals with stressful environmental conditions.

Figure 3- Expression profile of YGR058W and similar genes when exposed to low phosphate concentrations
This led me to look at how YGR058W's expression profile changes with different environmental stresses. The results of this search were mixed. The gene was often repressed, not induced, under stressful conditions. Heat and cold shock produced mild repression (1 -> 1.5 fold) as did exposure to DIT. Yet menadione expsure, nitrogen depletion, and sorbital exposure produced mild induction (again 1 -> 1.5 fold). These findings are difficult to interpret because they are contradictory. Equally confounding is the fact that YGR058W showed induction during part of the yeast stationary phase but repression during other parts. Why would the expression of this gene change from induction to repression hourly and over the first few days (Figure 4)? There is a trend of induction for the first few hours and then repression after a couple days that was repeated in two experiments. It's not clear what this finding means.
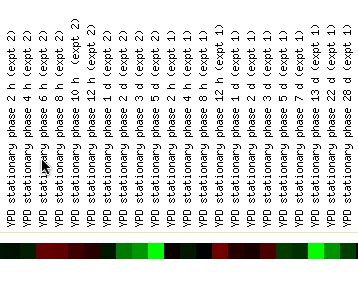
Figure 4 - YGR058W's expression profile during stationary phase
The microarray data from the expression during sporulation test was far more promising. The microarray data showed that as the sporulation process progressed YGR058W went from normal transcription to induction of around 1.5 fold at 11.5 hours. Thus it appears that the gene is activated during sporulation. Sporulation is a process that yeast undergo to form halpoid cells from diploid cells. This involves meiosis and spore morphogenesis (Details).

Figure 5 - YGR058W expression profile during sporulation
Further, when I looked at genes with similar expression profiles I found several genes that were involved in functions relating in some way to proliferation and cell replication. One gene SSP2 was involved in spore wall formation while another, PRM6, was involved in cellular fusion, and another, CLB6, was involved inthe transition of the mitotic cell cycle. The connection between any of these genes is not clear but the fact that genes involved in similar processes and with similar functions continue to reappear near each other means that we are narrowing down the function of this gene.
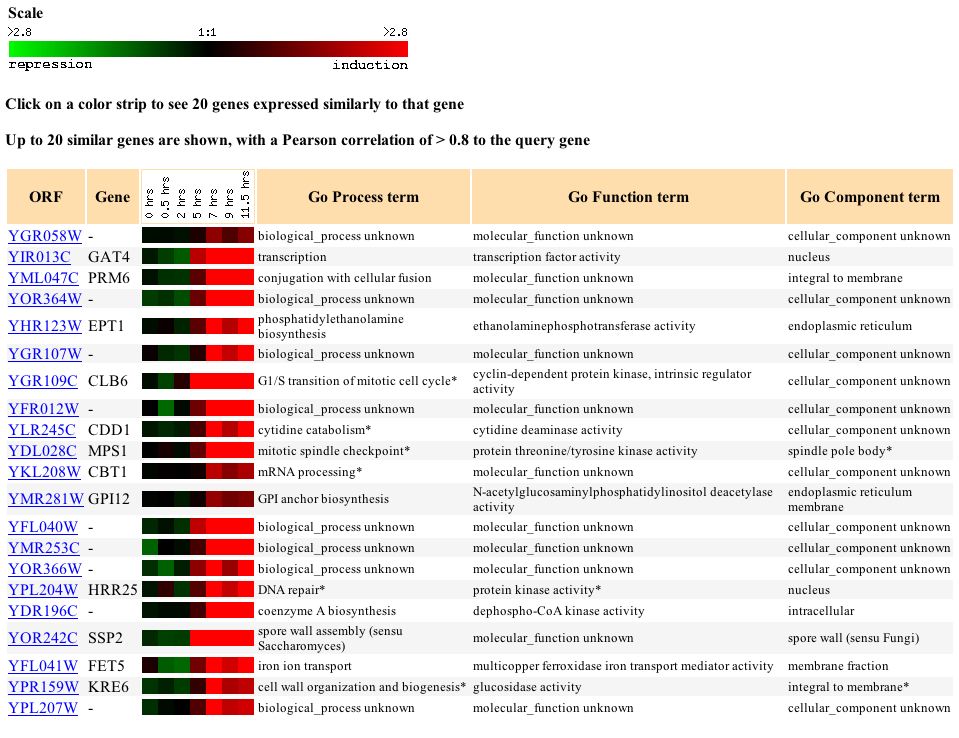 Figure 6 - Genes
with similar expression profiles to YGR058W during sporulation
Figure 6 - Genes
with similar expression profiles to YGR058W during sporulation
Searching Function Junction I found three conditions under which YGR058W was greatly induced (Figure . When exposed to griseofulvin, an antifungal agent, the yeast cell induced YGR058W transcription greatly (up to around 3 fold). If this gene was involved in the sporulation process this great induction would make sense. It is conceivable that the yeast cell, under the stress of a toxic agent, would shift toward sporulation and away from sexual reproduction for survival.
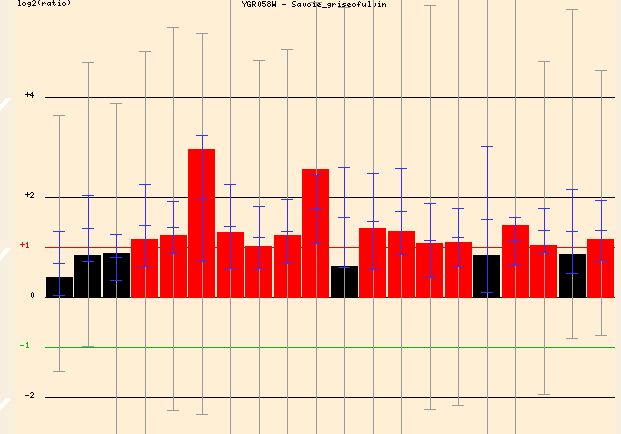
Figure 7 - The induction of YGR058W in response to increasing griseofulvin concentration. Graph is split into different concentrations of the agent. The general trends though are similar regardless of agent concentration.
The expression profile of YGR058W when exposed to salt yields similar findings. The high salinity results in a 1.5 to 2.5 fold induction of YGR058W after 30 and 90 minutes of exposure. This again makes sense if YGR058W is somehow related to the sporulation process. The stress of a high salinity environment would make sporulation a good option for yeast cells attempting to insure survival.
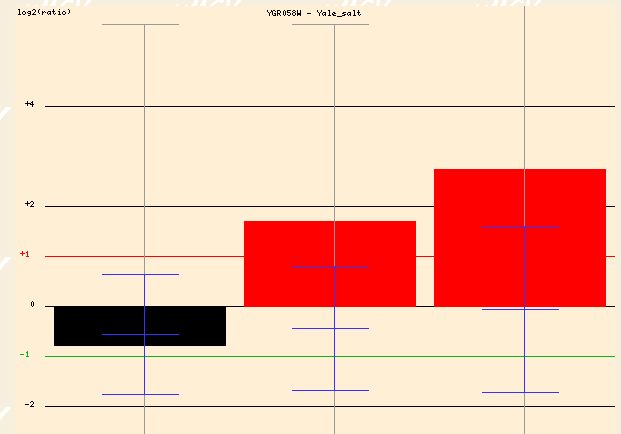
Figure 8 - Expression profile of YGR058W in response to high salinity at 0, 30, and 90 minutes)
Function Junction had an elaborate tree of genes that interact with YGR058W. Unfortuantely, most of these genes are also not annotated yielding little information. HOG1 is a gene involved in the hyperosmotic response. It's closeness to YGR058W makes it possible that their functions are similar. RCK1 is involved in the regulation of meiosis, an important part of sporulation. This supports the finding that YGR058W is somehow involved in this process. All of my findings continue to point with a somewhat vague certainty to YGR058W involvement in sporulation. Further research is necessary to uncover its exact role.
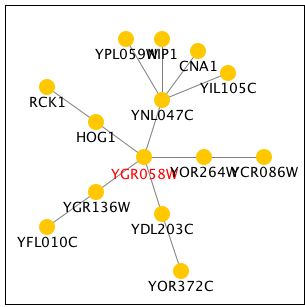
Figure 9 - Tree of genes that interact with YGR058W
Citations:
Expression Connection - http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl
Function Junction -http://db.yeastgenome.org/cgi-bin/SGD/functionJunction
Contact: Alex Trzebucki