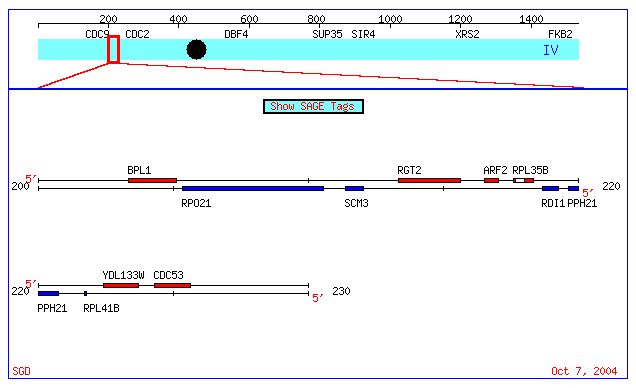
Figure 1. Chromosome location of RGT2 and YDL133W. Both genes are located on Chromosome 4 in the S. Cerevisiae genome. Image obtained from SGD database.
My Favorite Yeast Genes
Introduction:
The purpose of this assignment was to study two yeast (S. Cerevisiae) genes, one annotated and one unannotated. I chose the genes RGT2 (annotated) and the unannotated YDL133W. After gathering as much information about RGT2, I used the same databases (NCBI, BLAST, SGD, etc.) to try and find out as much information as possible about the unannotated gene YDL133W. My findings for both genes are found below.
RGT2:
RGT2 is a gene that can be found on chromosome 4 of the S. Cerevisiae genome (see figure below). A search of the S. Cerevisiae genome database returned the following results.

Figure 1. Chromosome location of RGT2 and YDL133W. Both genes
are located on Chromosome 4 in the S. Cerevisiae genome. Image obtained
from SGD
database.
Biologcial Process:
RGT2 is involved in signal transduction as well as response to glucose stimulus.
Molecular Function:
RGT2 is important for glucose binding. The protein product of this gene is a glucose receptor. As such, RGT2 is also important in receptor activity.
Cellular Component:
RGT2 can be found on the plasma membrane of S. Cerevisiae. A Kyte-Doolittle Hydropathy Plot confirms that there are transmembrane portions of the protein encoded by RGT2 (see figure below).
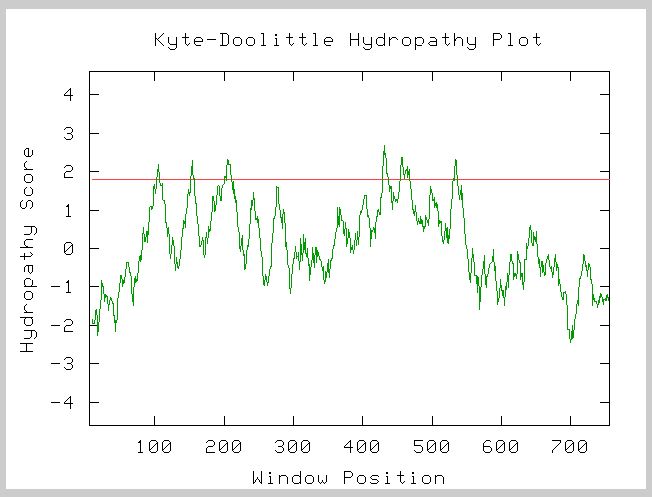
Figure 2. A Kyte-Doolittle Hydropathy Plot of RGT2. Window size
= 19. Peaks above 1.8 (red line) indicate a transmembrane component. RGT2
has 6 peaks above 1.8 indicating that it is a transmembrane protein.
Additional Information about RGT2:
The accession number for the entire chromosome 4 of S. Cerevisiae is NC_001136
Mutant Strains:
Several mutant strains of RGT2 have been found. All of these mutants relate to the growth of yeast with the mutant RGT2 (see figure below).
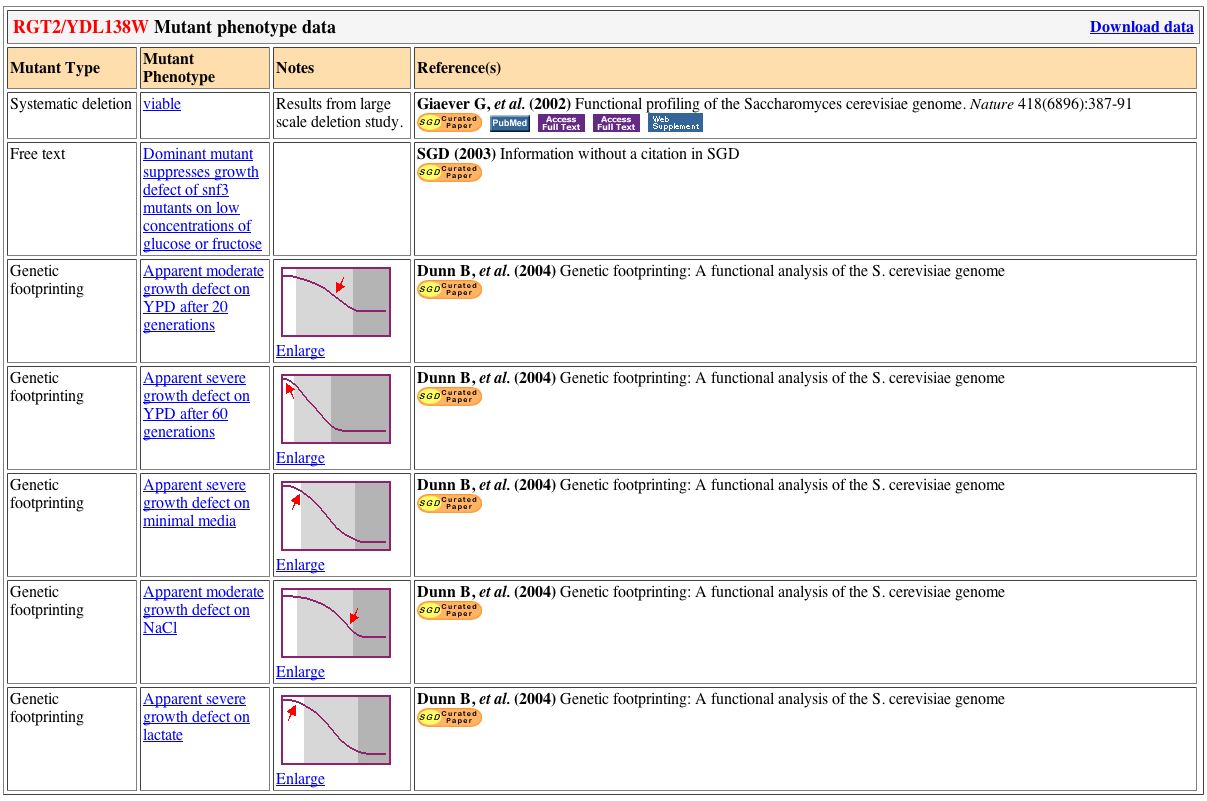
Figure 3. Mutant Phenotypes of RGT2. All of the mutant phenotypes
impact the growth rate of yeast. Image obtained from SGD.
Sequence Information
Genomic DNA sequence of RGT2:
ATGAACGATAGCCAAAACTGCCTACGACAGAGGGAAGAAAATAGTCATCTGAATCCTGGA
AATGACTTCGGCCACCACCAGGGTGCAGAATGTACGATAAATCATAACAACATGCCACAC
CGCAATGCATACACAGAATCTACGAATGACACGGAAGCAAAGTCCATAGTGATGTGCGAC
GATCCTAACGCATACCAAATTTCCTACACAAATAATGAGCCGGCGGGAGATGGAGCTATA
GAAACCACGTCCATTCTACTATCGCAACCGCTGCCGCTGCGATCGAATGTGATGTCTGTC
TTGGTAGGCATATTTGTTGCCGTGGGGGGCTTCTTGTTTGGGTATGACACTGGACTTATA
AACAGTATCACGGATATGCCGTATGTTAAAACCTACATTGCTCCGAACCATTCATATTTC
ACCACTAGCCAAATAGCCATACTCGTATCATTCCTCTCCCTAGGAACATTTTTCGGTGCG
TTAATCGCTCCCTATATTTCAGATTCATATGGTAGGAAGCCAACAATTATGTTTAGTACC
GCTGTTATCTTTTCCATCGGAAACTCATTACAGGTGGCATCCGGTGGCTTGGTGCTATTA
ATCGTCGGAAGAGTGATCTCAGGTATCGGGATCGGGATAATCTCTGCTGTGGTTCCTCTT
TATCAAGCTGAAGCTGCGCAGAAGAACCTTAGAGGTGCCATCATTTCCAGTTATCAGTGG
GCTATCACTATTGGGTTACTCGTGTCCAGTGCAGTATCGCAAGGAACTCATTCCAAAAAT
GGCCCGTCTTCATATAGAATACCAATTGGTTTGCAGTACGTTTGGTCAAGTATTTTAGCT
GTGGGCATGATATTCCTTCCAGAGAGTCCAAGATATTACGTCTTGAAGGATGAACTCAAT
AAAGCTGCAAAATCGTTATCCTTTTTAAGAGGCCTCCCGATCGAAGATCCAAGACTCTTA
GAGGAGCTTGTTGAAATAAAAGCCACTTACGATTATGAAGCATCGTTCGGCCCGTCAACA
CTTTTAGATTGTTTCAAAACAAGTGAAAATAGACCCAAACAGATTTTACGAATATTTACT
GGTATCGCCATACAAGCTTTTCAACAGGCATCTGGTATCAATTTTATATTCTACTATGGA
GTTAATTTTTTCAACAACACAGGGGTGGACAACTCTTACTTGGTTTCTTTTATCAGCTAT
GCCGTCAACGTCGCCTTCAGTATACCGGGTATGTATTTAGTGGATCGAATTGGTAGAAGA
CCAGTCCTTCTTGCTGGAGGTGTCATAATGGCAATAGCAAATTTAGTCATTGCCATCGTT
GGTGTTTCCGAGGGAAAAACTGTTGTTGCTAGTAAAATTATGATTGCTTTTATATGCCTT
TTCATTGCTGCATTTTCGGCGACATGGGGTGGTGTCGTGTGGGTGGTATCTGCTGAACTG
TACCCACTTGGTGTCAGATCGAAATGTACCGCCATATGCGCTGCCGCAAATTGGCTAGTT
AATTTCACCTGTGCCCTGATTACACCTTACATTGTTGATGTCGGATCACACACTTCTTCA
ATGGGGCCCAAAATATTCTTCATTTGGGGCGGCTTAAATGTCGTGGCCGTTATCGTTGTT
TATTTCGCTGTTTATGAAACGAGGGGATTGACTTTGGAAGAGATTGACGAGTTATTTAGA
AAGGCCCCAAATAGCGTCATTTCTAGCAAATGGAACAAAAAAATAAGGAAAAGGTGCTTA
GCCTTTCCCATTTCACAACAAATAGAGATGAAAACTAATATCAAGAACGCTGGAAAGTTG
GACAACAACAACAGTCCAATTGTACAGGATGACAGCCACAACATAATCGATGTGGATGGA
TTCTTGGAGAACCAAATACAGTCCAATGATCATATGATTGCGGCGGATAAAGGAAGTGGC
TCGTTAGTAAACATCATCGATACTGCCCCCCTAACATCTACAGAGTTTAAACCCGTGGAA
CATCCGCCAGTAAATTACGTCGACTTGGGGAATGGTTTGGGTCTGAATACATACAATAGA
GGTCCTCCTTCTATCATTTCTGACTCTACTGATGAGTTCTATGAGGAAAATGACTCTTCT
TATTACAATAACAACACTGAACGAAATGGAGCTAACAGCGTCAATACATATATGGCTCAA
CTAATCAATAGCTCATCTACTACAAGCAACGACACATCGTTCTCTCCATCACACAATAGC
AATGCAAGAACGTCCTCTAATTGGACGAGTGACCTCGCTAGTAAGCACAGCCAATACACT
TCCCCCCAATAA
Amino acid sequence of RGT2:
MNDSQNCLRQREENSHLNPGNDFGHHQGAECTINHNNMPHRNAYTESTNDTEAKSIVMCD
DPNAYQISYTNNEPAGDGAIETTSILLSQPLPLRSNVMSVLVGIFVAVGGFLFGYDTGLI
NSITDMPYVKTYIAPNHSYFTTSQIAILVSFLSLGTFFGALIAPYISDSYGRKPTIMFST
AVIFSIGNSLQVASGGLVLLIVGRVISGIGIGIISAVVPLYQAEAAQKNLRGAIISSYQW
AITIGLLVSSAVSQGTHSKNGPSSYRIPIGLQYVWSSILAVGMIFLPESPRYYVLKDELN
KAAKSLSFLRGLPIEDPRLLEELVEIKATYDYEASFGPSTLLDCFKTSENRPKQILRIFT
GIAIQAFQQASGINFIFYYGVNFFNNTGVDNSYLVSFISYAVNVAFSIPGMYLVDRIGRR
PVLLAGGVIMAIANLVIAIVGVSEGKTVVASKIMIAFICLFIAAFSATWGGVVWVVSAEL
YPLGVRSKCTAICAAANWLVNFTCALITPYIVDVGSHTSSMGPKIFFIWGGLNVVAVIVV
YFAVYETRGLTLEEIDELFRKAPNSVISSKWNKKIRKRCLAFPISQQIEMKTNIKNAGKL
DNNNSPIVQDDSHNIIDVDGFLENQIQSNDHMIAADKGSGSLVNIIDTAPLTSTEFKPVE
HPPVNYVDLGNGLGLNTYNRGPPSIISDSTDEFYEENDSSYYNNNTERNGANSVNTYMAQ
LINSSSTTSNDTSFSPSHNSNARTSSNWTSDLASKHSQYTSPQ*
An NCBI BLASTn search using the above genomic DNA sequence returned the following results for RGT2.
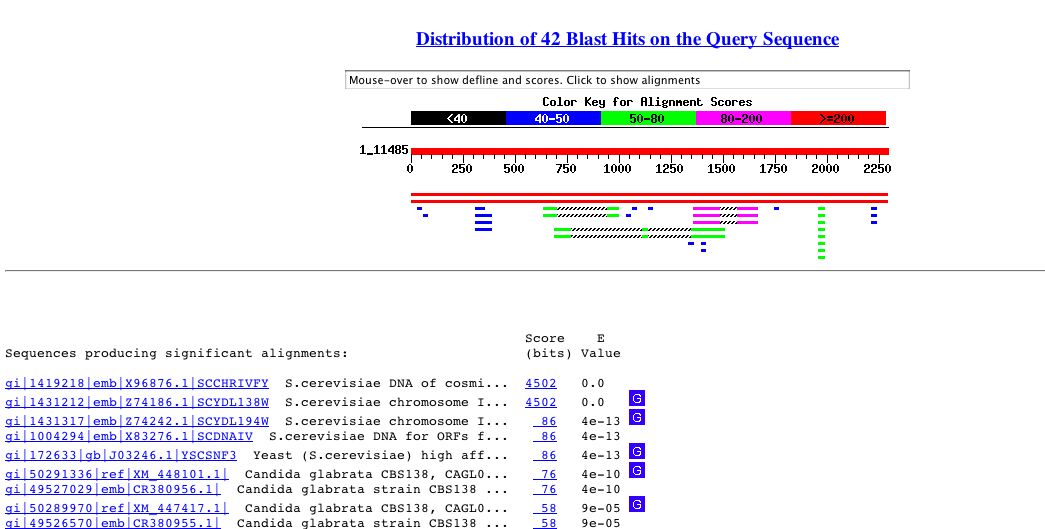
Figure 4: Results of BLASTn search using RGT2 genomic DNA sequence.
Two hits were obtained that were identical to the sequence I entered. These
hits were from the complete genome of chromosome 4 and the specific gene
in question, RGT2. Several other hits had high E-values including another
yeast gene involved in glucose transport (SNF3) and several genes from
Cadida glabrata.
A HomoloGene search of RGT2 returned the following results:

Figure 5. Results of HomoloGene search for RGT2. There are four species in which this gene is conserved. For more information click here.
Further information about RGT2 can be found at Entrez
Gene.
YDL133W:
According to the S. Cerevisiae database there are no known biological processes in which YDL133W is involved. Furthermore, there are no molecular functions of cellular components that can be attributed to this gene. To see more information go to SGD.
Additional Information about YDL133W:
Mutant Strains:
Only two mutant strains of YDL133W exist. When deleted from the yeast genome, the yeast remains viable in one case, but is sensitive to pH8 after 15 generations.
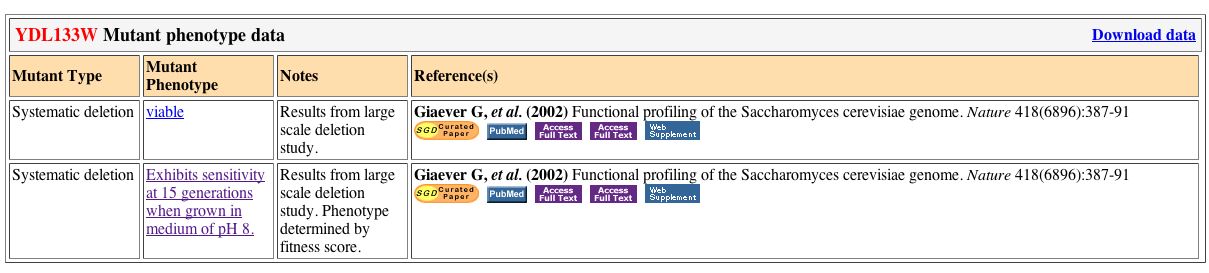
Figure 6. Mutant Phenotype Data for YDL133W. Obtained from
SGD.
Sequence Information:
The genomic DNA sequence of YDL133W is:
ATGGGGGATTCTAATTCCAGCCAAGAAGCGTATTCTGATACAACATCCACGAACGCTTCT
AGAATCGCAGATCAAAATCAACTTAATTTAAACGTGGATCTTGAAAAAAATCAAACAGTA
AGGAAGTCAGGTTCACTGGAAGCATTACAGAATGCAAAGATACATGTTCCCAAACACAGT
GATGGTTCACCACTAGACTATCCAAAATTAAATACTTACACGTTTGTGCCTACCACAGTG
CCGCCTTATGTTTTAGAGGCGCAGTTTGATAAATTAAGACTTCAGGATAAGGGCACCGTT
GATGGAAATGTTACCGATGATAAAAATCTTCCAAAAGAGTTTAAATGGGGCCAGTTTGCA
TCTACCATCGGCTGTCATTCTGCATACACTAGAGACCAAAACTACAATCCTAGTCACAAA
TCCTACGATGGTTACTCTCTATCATCATCTACCTCATCAAAAAATGCGGCTCTTAGAGAA
ATTTTAGGTGACATGTGCAGCGAGTGGGGAGGTGAAGAACGACTAGAAGGTGTTCTACAT
TCAGAAATAGGTGCCAATTTAGAGTTCAACACGACAGAAGAGAGAAAAGAGTGGTTACAA
TATATCGAAAAGGTAAAAGATTTCTATTACGGTGATAATAAGAAGAACCCAGAATCACCC
GAATCAGTACACAACAAAGTTTACAAGTCAGATTGGGTAAATGAGCTCAATAAAGAGAGA
GAAAAGTGGCGGAGGTTAAAGCAAAGGAAGTTACAACAGTGGAGACCTCCTTTGACGTCG
CTATTACTTGATAATCAATATTTGATTTTGGGTTTGAGGATTTTCACTGGTATATTATCG
TGCATTTCACTTGCTCTGGCAATCAAGATTTTTCAAAACTCAAGATCAAATAACACCATC
AGTGAAAGTAAAATCGGGCAACAACCAAGCACCATAATGGCCATCTGCGTCAACGCCGTG
GCGATTGCGTATATTATTTACATTGCACACGATGAATTTGCTGGTAAACCTGTTGGTTTG
AGGAATCCGTTAAGCAAACTGAAACTAATTTTGTTGGACTTACTTTTTATAATTTTTTCA
AGCGCTAATCTAGCGCTGGCATTTAATACTCGTTTCGACAAAGAGTGGGTTTGCACTTCT
ATTCGCAGGTCAAATGGAAGCACGTATGGATACCCAAAAATACCGCGTATCTGCAGAAAA
CAGGAAGCTTTATCCGCTTTTCTGTTCGTTGCCTTGTTTATGTGGGTAATCACTTTCTCG
ATCAGTATTGTTAGAGTAGTCGAAAAAGTCAGTTCAATTACCAACAGAAACTGA
The amino acid sequence of YDL133W is:
MGDSNSSQEAYSDTTSTNASRIADQNQLNLNVDLEKNQTVRKSGSLEALQNAKIHVPKHS
DGSPLDYPKLNTYTFVPTTVPPYVLEAQFDKLRLQDKGTVDGNVTDDKNLPKEFKWGQFA
STIGCHSAYTRDQNYNPSHKSYDGYSLSSSTSSKNAALREILGDMCSEWGGEERLEGVLH
SEIGANLEFNTTEERKEWLQYIEKVKDFYYGDNKKNPESPESVHNKVYKSDWVNELNKER
EKWRRLKQRKLQQWRPPLTSLLLDNQYLILGLRIFTGILSCISLALAIKIFQNSRSNNTI
SESKIGQQPSTIMAICVNAVAIAYIIYIAHDEFAGKPVGLRNPLSKLKLILLDLLFIIFS
SANLALAFNTRFDKEWVCTSIRRSNGSTYGYPKIPRICRKQEALSAFLFVALFMWVITFS
ISIVRVVEKVSSITNRN*
Using the above genomic DNA sequence a BLASTn search was performed. I obtained the following results:
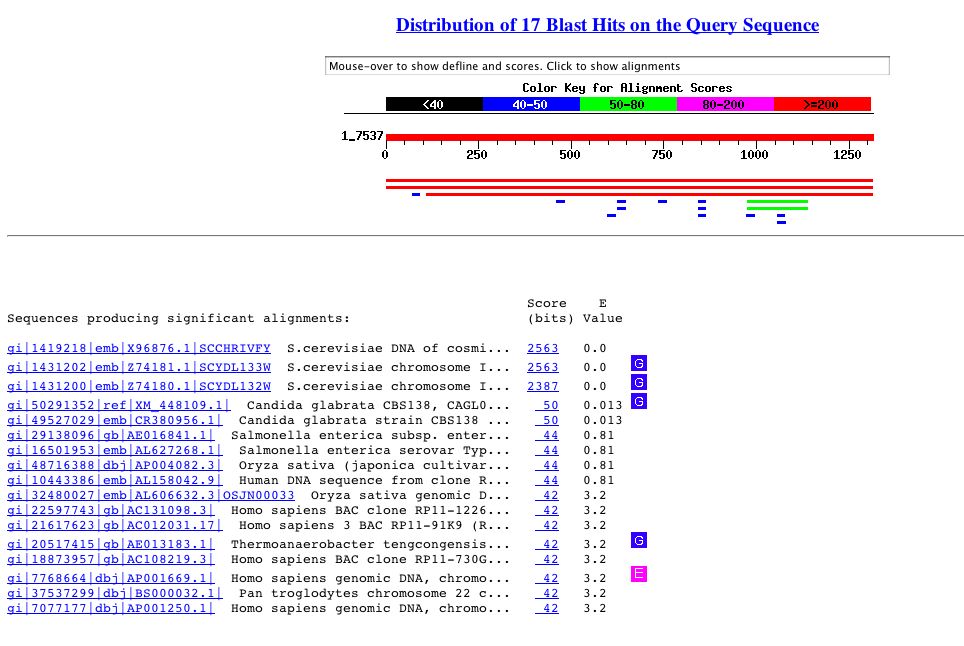
Figure 7. BLASTn search of the genomic DNA sequence of YDL133W.
The only hits that are simialr (i.e. low E values) to the YDL133W gene
are part of the chromosome 4 genome and other chromosome 4 sections. Image
obtained from here.
A search of Entrez Gene stated that YDL133W is a probable membrane protein. The link to this information can be found here.
A Kyte-Doolittle Hydropathy Plot of the amino acid sequence of YDL133W confirmed the fact that it is probably a membrane protein.
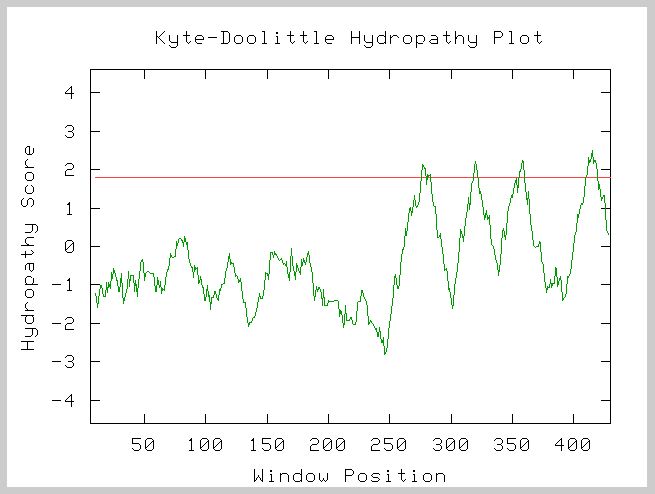
Figure 8. Kyte-Doolittle Hydropathy Plot for the amino acid sequence of YDL133W. Window size = 19. Peaks above 1.8 (red line) indicate transmembrane components of the protein. YDL133W has four peaks above 1.8 suggesting that it is a transmembrane component of the cell.
A HomoloGene search of YDL133W returned the following results:

Figure 9. HomoloGene search results for YDL133W. The only homologous
gene is found in E. Gossypii. For more information about the homolog
click
here.
Further information about YDL133W can be found at Entrez
Gene.
Conclusion of Cellular Role:
Based on the information gathered from the Kyte-Doolittle Hydropathy
Plot I conclude that YDL133W is a transmembrane protein. In addition, based
on the mutant phenotypes of S. Cerevisiae and the sensitivity at
pH 8 after 15 generations when YDL133W is deleted, I hypothesize that YDL133W
is involved in cellular regulation in varying pH ranges.
References:
Kyte-Doolittle Hydropathy Plot. 2004. <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm> Accessed 7 October 2004.
NCBI. 2004. <http://www.ncbi.nlm.nih.gov/>
Accessed 7 October 2004.
Saccharomyces Genome Database. 2004. <http://www.yeastgenome.org/>
Accessed 7 October 2004.