This web page was produced as an assignment for an undergraduate course at Davidson College
My Favorite Yeast Genes: ASP3-4 (Annotated) and YLR161W (Non-annotated)
These two genes ASP3-4 and YLR161W are found in the genome of yeast, Saccharomyces cerevisiae. While ASP3-4 is annotated and its gene ontology (biological process, molecular function and cellular component) is known, the non-annotated gene, YLR161W, is an ORF (open reading frame) found slightly upstream of ASP3-4 and has no information with regards to it's gene ontology. I will present the information on the annotated gene, ASP3-4, and then try to figure out the non-annotated gene's, YLR161W's, role in the life of yeast through sequence analysis.

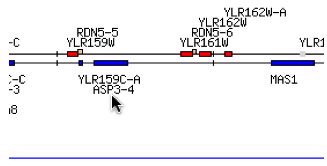
Fig. 1- Location of ASP3-4 and YLR161W on chromosome XII of yeast. Image used courtesy of http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004150
General information for ASP3-4:
ASP3-4 is one of four genes of ASP3 found in the genome of yeast. This gene produces the protein cell-wall L-asparaginase II. This enzyme catabolizes asparagine and in environments with low levels of nitrogen, the expression of this gene occurs. The overall function of the gene is to create a nitrogen source for yeast in nitrogen deprived environments.
Molecular Function:
Molecular function refers to what the gene product actually does (and is not the gene product itself), like whether it is a protein that binds to something or an enzyme that catalyzes a reaction. As for ASP3-4 its gene product is an enzyme and its molecualr function is the catalysis of a reaction. Cell-wall L-asparaginase is a catalyst in the reaction: L-asparagine + H2O = L-aspartate + NH3.
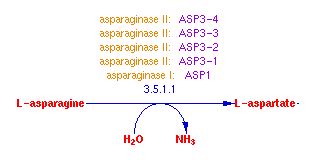
Fig. 2- A diagram of the molecular function of ASP3-4. Image used courtesy: http://pathway.yeastgenome.org:8555/YEAST/new-image?type=PATHWAY&object=ASPARAGINE-DEG2-PWY&detail-level=2
Biological Process:
Several molecular functions contribute to an overall biological process. ASP3-4's biological process is the breakdown of asparagine into 2-amino-3-carbamoylpropanoic acid. Also, only in nitrogen deprived environments, does the expression of ASP3-4 occur (This means that ASP3-4 is involved in the biological process of cellular response to low levels of nitrogen).
Cellular Component:
Cellular component refers to where in the cell the activity of the gene product occurs. As for ASP3-4, the cellular component is the periplasmic space, the area between the plasma membrane and the cell wall.
ASP3-4's interaction with other genes and their gene products:
The formation of L-asparaginase II is dependent upon the gene GLN3 and its response to low levels of nitrogen is dependent upon URE2's gene product.
Structure of L-asparaginase II:
Fig. 3- A picture of the protein, L-asparaginase II (actually, it's an homolog of a similar protein in another organism). Image used courtesy http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics;pdbId=1hfw&opt=show&size=500
Why study ASP3-4?
Because the production of L-asparaginase is regulated by the levels of nitrogen available, it can be used as a model in the study of secreted proteins in yeast. Although the enzyme activity and protein interactions in the periplasmic space appear to be regulated/imduced by the levels of nitrogen present, the actual mechanisms for this regulation/induction are not well understood. In a recent study to figure out this mechanism, a group of researchers made a chimera gene(ASP-GFp) that produced asparaginase tagged with a green fluorescent protein to study the dynamics of protein secretion in vivo.
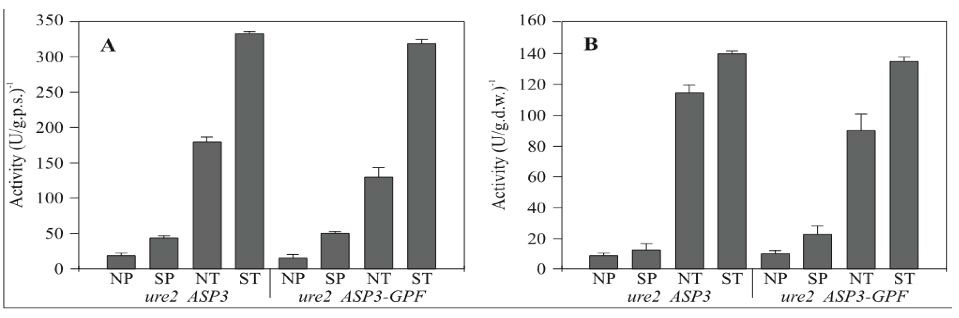
Fig. 4- These are two graphs from the ASP-GFP gene study. The graphs represent Asparaginase II activity levels on the y-axis and the symbols on the x-axis represent (N) non-nitrogen starved, (S) nitrogen starved, (P) periplasmic activity, and (T) total enzymatic activity. (A) wild type. (B) URE2 strain. Permission pending on use of image.
YLR161W: Sequence Analysis and Predicted Function
Sequence Analysis:
I ran a BLASTn on the the nucleotide sequence of YLR161W and I got several hits for matches in the yeast genome sequence as expected. An unexpected result from this blast is one of the hits describes the sequence as a 5S rRNA gene and its flanking sequence. So, perhaps this potential gene is involved in the rRNA somehow.
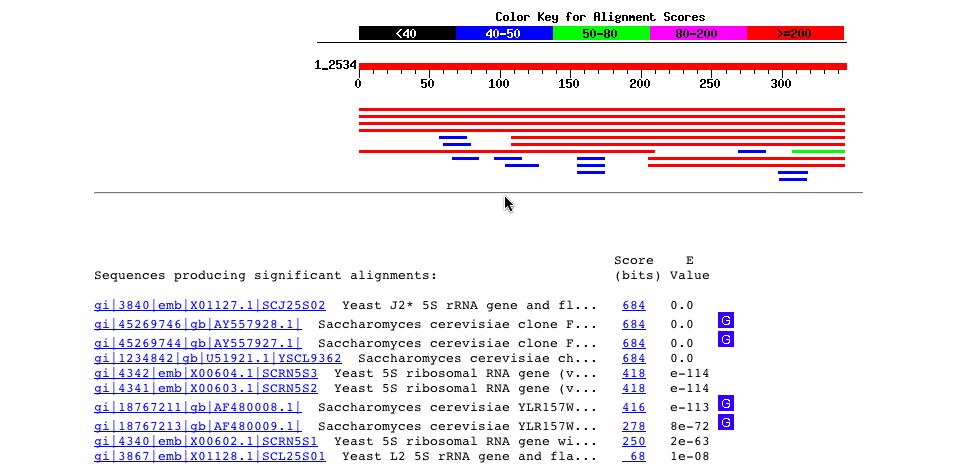
Fig. 5 - A picture of the blastn results for the nucleotide sequence of YLR161W. Image used courtesy http://www.ncbi.nlm.nih.gov:80/BLAST/Blast.cgi
The BLASTp of the amino acid sequence gave no significant results.
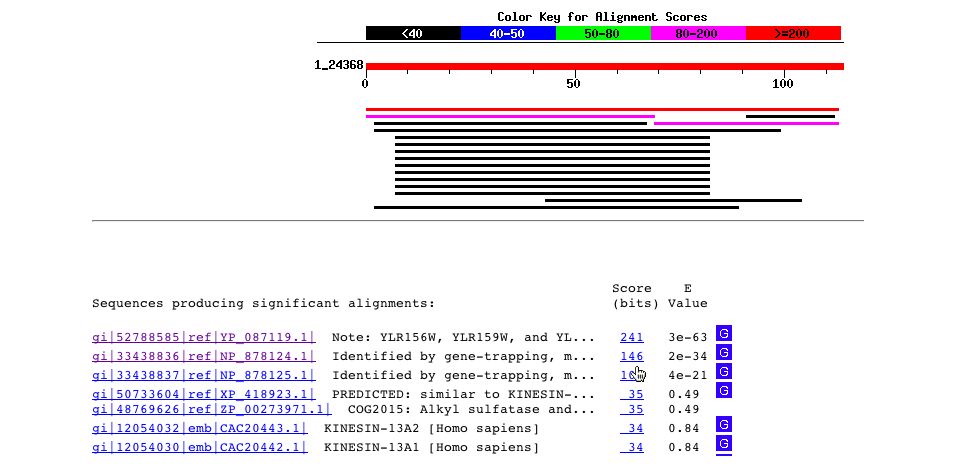
Fig. 6- A blastp search for the amino acid sequence just gives hits for the sequence itself in yeast and the sequence is not found in any other organisms. The sequence found in Homo sapiens has such a high e-value that it can be ignored. Image courtesy http://www.ncbi.nlm.nih.gov:80/BLAST/Blast.cgi
A conserved domain search also produced no results. A Kyte-Doolittle analysis of the amino acid sequence gave this graph:

Fig. 7- A Kyte -Doolittle graph shows that the amino acid sequence transcribed by YLR161W does not produce a transmembrane protein. Image used courtesy http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/slrkd.pl
Entering the amino acid sequence into the PREDATOR database yields the following:
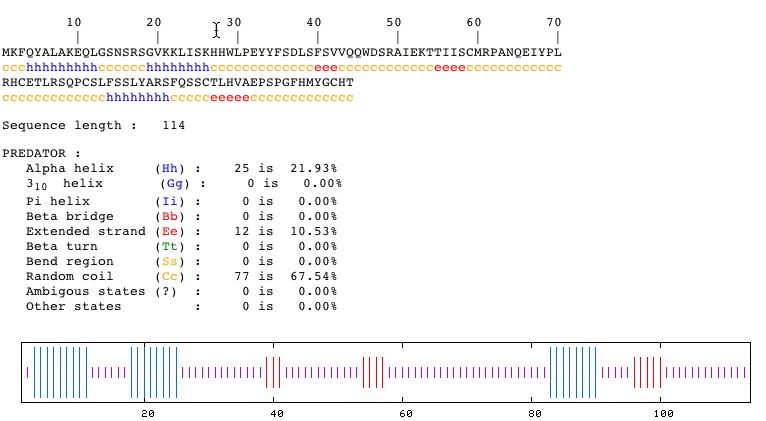
Fig. 8- Secondary structure of the protein coded by YLR161W. Image courtesy http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl
This picture from PREDATOR shows that the secondary structure of the protein has three alpha helices and three beta bridges.
Predicted Function:
Overall, there is not much data for this particular sequence (no orthologous genes or conserved domains in the protein). I predict that this gene is involved in the rRNA of yeast.
References:
1) Bon EP, et al.1997. Asparaginase II of Saccharomyces cerevisiae. GLN3/URE2 regulation of a periplasmic enzyme. Appl Biochem Biotechnol 63-
65():203-12
2) Sinclair K, et al. 1994. The ASP1 gene of Saccharomyces cerevisiae, encoding the intracellular isozyme of L-asparaginase. Gene 144(1):37-43
3) Gene Ontology Software Group. 2004. Gene Ontology Database.<http://www.godatabase.org/dev/database/> Accessed 2004 7 Oct.
4) NCBI. 2003. National Center for Biotechnology Information. <http://www.ncbi.nih.gov/> Accessed 2004 7 Oct.
5) Dolinski, K. et. al. 2004. Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 2004 7 Oct.
6) Martins, A.S. et. al. 2003. AsparaginaseII-GFP Fusion as a Tool for Studying the Secretion of the Enzyme under Nitrogen Starvation.
Brazilian Journal of Microbiology 34:373-377.
Questions or comments? email me: jobunton@davidson.edu
John Bunton's Genomics Home Page