This web page was produced as an assignment for an undergraduate course at Davidson College. My Favorite Yeast Proteins: Cox17 and YLL007C |
Introduction: For this assignment I searched various protein databases to confirm my earlier findings regarding the annotated yeast gene Cox17 and the non-annotated hypothetical ORF YLL007C. As indicated previously, Cox17 and YLL007C are both located on yeast chromosome XII. Cox17 is known to function as a copper metallochaperone that transports copper from the cytosol to the mitochondrial inter-membrane space for delivery to cytochrome c oxidase. In the previous assignment, I predicted that the hypothetical ORF YLL007C is a nuclear membrane protein and is involved in nuclear migration during mitosis and mating. Since this prediction was made with limited information, it is important to investigate the role of YLL007C further using the available information from various protein databases. For clarity purposes, my own original thoughts reagrding the information that is presented in this website will appear in yellow shaded boxes throughout this page.
|
Cox17 Protein Information: Information from The Protein Data Bank: To find protein structure of Cox17, I began by searching the Protein Data Bank, the single worldwide repository for the processing and distribution of 3-D biological macromolecular structure data. I was able to locate a chime image of Cox17 bound to a copper ion. This image can be seen below: |
| |
| Figure 1: Chime image of Cox17 obtained from the Protein Data Bank (PDB ID: 1u96). It is important to note that this image is an NMR structure, depicting Cox17 in 20 different orientations and highlighting the binding site of the copper ion (maroon). Although it appears as if several copper ions are bound to the Cox17 protein, each ion is representative of a different orientation of Cox17, as the protein is not capable of binding 20 copper ions simultaneously. Alpha helices are highlighted in pink. |
Information from PROWL: Using the PROWL database, I was able to detemine the molecular mass and isoelectric point of the Cox17 protein. The database indicated that Cox17 is 8050.601 Da and has a pI of 4.9. |
Information from MIPS: Next, I decided to search the MIPS (Munich Information Center for Protein Sequences) database. Much of the information that I found on this page regarding Cox17 function was identical to the information that I found in the previous assignment. In addition to this knowledge, the MIPS database also proivides protein-protein interaction information. The data for Cox17 yielded the following conclusions: |
|
| Figure 2: Snapshot of an image obtained from a MIPS databse search for Cox17. |
| The MIPS database indicates that Cox17 interacts physically with JSN1 and genetically with SCO1 and SCO2. A physical interaction simply means that the proteins bind to one another within the cell. Genetic interactions often reveal redundancy in the genome, as genetic interactions are determined by mutant phenotypes that are able to be rescued by overexpression of other genes in the genome. In this case, it is known that the cytochrome oxidase deficiency seen in Cox17 mutants can be partially rescued by SCO1 when it is present on a high copy plasmid (Beers et al. 1997). With this information, I decided to determine whether or not these proteins that are both phisical and genetic interactions with Cox17 have functional similarities by performing a search using the Saccharomyces Genome Database. The results can be seen below: |
JSN1:
SCO1:
SCO2:
|
| Figure 3: Snapshots of information regarding JSN1, SCO1 and SCO2 yeast genes obtained from the Saccharomyces Genome Database. |
Although JSN1 has no obvious functional similarity to Cox17, SCO1 and SCO2 are both implicated in mitochondrial transport and insertion of copper into the active site of cytochrome c oxidase. Research has indicated that it is still not clear whether or not SCO1 is a carrier that transports copper to the matrix of the mitochondria or whether it acts as a transferase that can add copper directly to cytochrome oxidase (Beers et al. 1997). Presumably, since SCO2 is very similar to SCO1, the predictions for its role are similar to those of SCO1. Although a concrete model of these genetic interactions has not yet been determined, it is clear that Cox17, SCO1 and SCO2 have some overlapping functional roles. It is interesting to note that JSN1 is a member of a family of RNA binding proteins that interacts with the mRNA encoding membrane-associated proteins. I found this to be a bit puzzling considering the fact that Cox17 is found to be localized to the cytoplasm and intermembrane space of the mitochondria. Despite this fact, it is highly likely that the Cox17 protein interacts in some way with the mitochondrial membrane in order to shuttle copper from the cytosol to the mitochrondria. In this sense, one might be able to consider Cox17 to be a membrane associated protein due to its interactions with the mitochondrial membrane. Thus, it is possible that JSN1 binds Cox17 mRNA. |
Information from DIP: Since the MIPS database was able to indentify three proteins that interact either genetically or physically with Cox17, I decided to search the Database of Interacting Proteins to determine whether or not the results would be consistent. The graphical representation obtained from DIP which depicts Cox17 and those proteins with which it physically interacts can be seen below: |
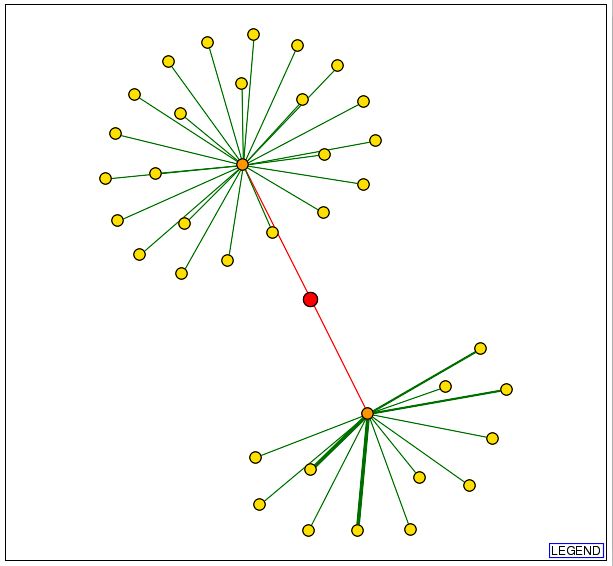  |
| Figure 4: Snapshot of image obtained from the Database of Interacting Proteins (DIP) using a search for Cox17. The red dot in the center of the image represents Cox17. The orange dot in the upper portion of the figure represents JSN1 and the orange dot in the lower portion of this figure represents Sec17. |
| This data indicates that there are two yeast proteins with which Cox17 physically interacts: JSN1 and Sec17. JSN1 was implicated in the previous search using the MIPS database. With this new information, I performed a search in the Saccharomyces Genome Database (SGD) to determine the function of the Sec17 protein. These results can be seen below: |
Sec17:
|
| Figure 5: Snapshot of image obtained from the Saccharomyces Genome Database performing a search for the Sec17 yeast gene. |
| Although Sec17 does not have a functional relationship to Cox17, it is interesting to note that Sec17 is involved in vesicular transport between the ER and Golgi. Perhaps that fact that Cox17 and Sec17 are both involved in intracellular transport promotes their physical interaction, however, from the data available the purpose of their physical interaction is not clear. It was surprising to me that that the DIP database did not indicate that Cox17 had any physical protein-protein interactions with any mitochondrial associated proteins, as Cox17 is thought to deliver copper to the intermembrane space of the mitochondria. Perhaps these physical interactions with mitochonrial proteins are not easily identified and thus, have not yet been confirmed. |
Information from SGD: In another effort to find protein-protein interactions between Cox17 and other proteins in the yeast genome, I searched the protein information section of the the Saccharomyces Genome Database (SGD). The SGD protein information contains results from yeast two hybrid assays performed by the Yeast Resouce Center. The results can be seen below: |
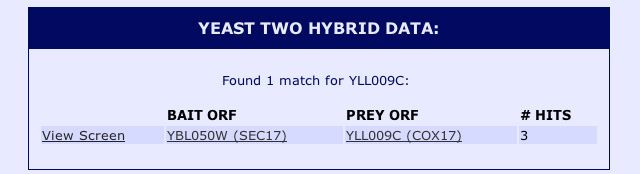 |
| Figure 6: Snapshot image of the yeast two hybrid data for Cox17 obtained from the Saccharomyces Genome Database protein information. |
The yeast two hybrid data confirmed the results from the DIP databse, as the results indicate that Cox17 interacts with Sec17 in yeast two hybrid experiments with YBL050W as the bait ORF. After searching these databases, it is clear that some databases (such as DIP and the Yeast Resource Center) were only able to identify proteins that physically interacted with Cox17. The proteins that physically interact with Cox17 do not seem to be functionally related, whereas those proteins that interact genetically with Cox17, such as SCO1 and SCO2, are both associated with the transport of copper to the mitochondria. It is also interesting to note that the other protein with which Cox17 is known to interact, JSN1, is not identified by the yeast two hybrid method. There are three possible explanations for this outcome. The most simple explanation is that JSN1 has not been tested by the yeast to hyrbid method as either a prey or a bait ORF, thus preventing the determination of its interacting proteins by this method. Another possible explanation is that JSN1 does not function well within the nucleus, and thus its interactions cannot be identified via the yeast two hyrbid method. The yeast two hybrid system works by fusing a bait ORF to a DNA binding domain and fusing a prey ORF to an activation domain of a complex required to initiate transcription. If the prey ORF and bait ORF interact, transcription of a reporter gene will be initiated. When proteins do not work well in the nuclues (i.e. they are misfolded) it is possible that they are prevented from binding proteins with which they normally interact. I would find it hard to believe that JSN1 does not function well in the nucleus given that it is an RNA binding protein, although it cannot be ruled out based upon the information provided. The third explanation is that JSN1 only binds the mRNA of Cox17, and since the yeast two hybrid method only identifies physical interactions between two mature protein products, the interaction between JSN1 and Cox17 could not be detected via this approach. Since JSN1 is known to be an RNA binding protein, it is my hypothesis that the third explanation is the most plausible. |
Information from KEGG : As the last part of my investigation of the Cox17 protein, I searched the KEGG database to identify the biochemical pathway in which Cox17 plays a role. The KEGG Pathway database is a collection of graphical diagrams (KEGG pathway maps) representing molecular interaction networks in various cellular processes. A search for Cox17 yielded the following results: |
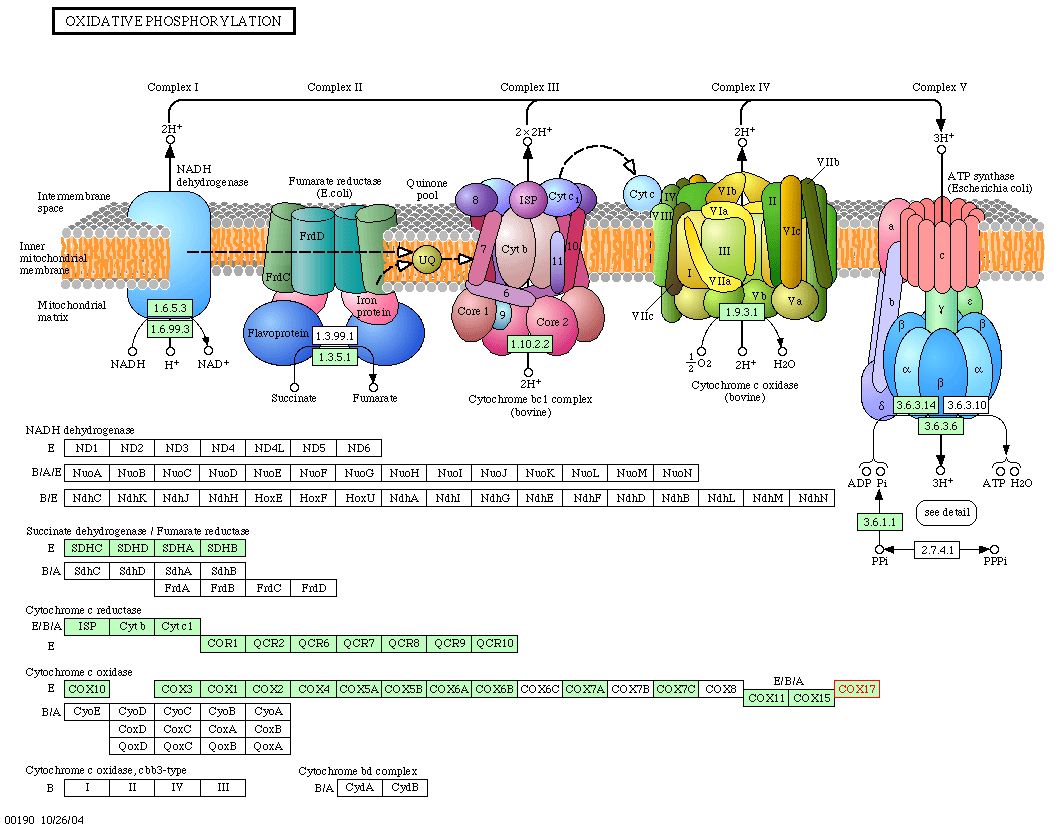 |
| Figure 7: Snapshot of image obtain from KEGG depicting the oxidative phosphorylation pathway in yeast. Cox17 is implicated in this pathway, as it is necessary for the proper assembly of cytochrome c oxidase. |
Information from PDF Files: No information regarding Cox17 was found in any of the PDF files (Aging, Benno Figure 1, Benno Figure 2, Degredation) that were discussed in class.
|
Conclusions: Although Cox17 has been fairly well characterized, there are a few additional experiments that could be done in order to have a more complete picture of this protein's cellular role. First, it would be useful to know if the JSN1 protein bound to Cox17 mRNA. The mature JSN1 protein should be used as the bait ORF in the yeast two hybrid method along with Cox17 as the prey ORF to rule out the possibility that these two proteins do not interact in their mature form. If no result was found from the yeast two hybrid study, it would be easier to make conjectures about how these two proteins interact. Also, it would be helpful to have more specific functional descriptions of SCO1 and SCO2. At this point, it appears as if SCO1 and SCO2 are potentially functionally equivalent, however, more investigation would have to be done to confirm this hypothesis. One way to make this determination would be to see if SCO2 present in a high copy plasmid was able to completely rescue a SCO1 null mutant and vise versa. Finally, it would be interesting to determine if any physical protein interactions could be determine between Cox17 and other mitochondrial associated proteins. Presumably, if Cox17 is delivering copper to cytochome c oxidase in the mitochondrial intermembrane space, it must have some physical interactions with mitochondrial membrane proteins. Once again, the yeast two hybrid method could be used to determine whether or not using mitochondrial associated proteins as bait ORF's were able to interact with the Cox17 protein as a prey ORF. It is important to note however that these interaction studies may have already been tested and simply not detected due to the inability of certain mitochonrdial proteins to work well in the nucleus. Overall, the Cox17 protein is fairly well characterized and all of the data that I found regarding the protein was consistent with the prediction that Cox17 is a copper metallochaperone that delivers copper to the inter membrane space of the mitochondria. |
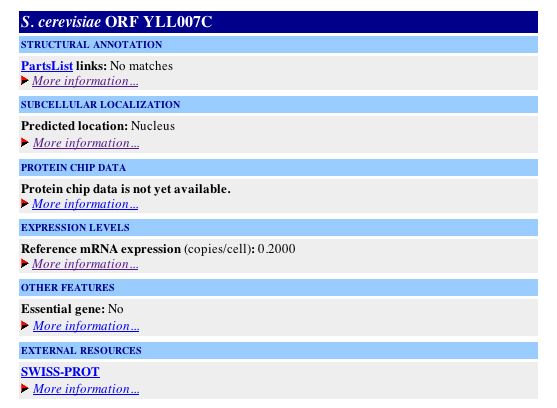
YLL007C Protein Information: Information from Protein Data Bank: A search of the Protein Data Bank yielded no results for YLL007C. Since the ORF has no known molecular function, it is not surprising that no structural data exists for this protein. Information from Yeast Microarray Global Viewer: Since I was unable to obtain structural information from PDB, I decided to search the Yale Info database via the Yeast Microarray Global Viewer to obtain the structural annotation for the YLL007C ORF. The results can be seen below: |
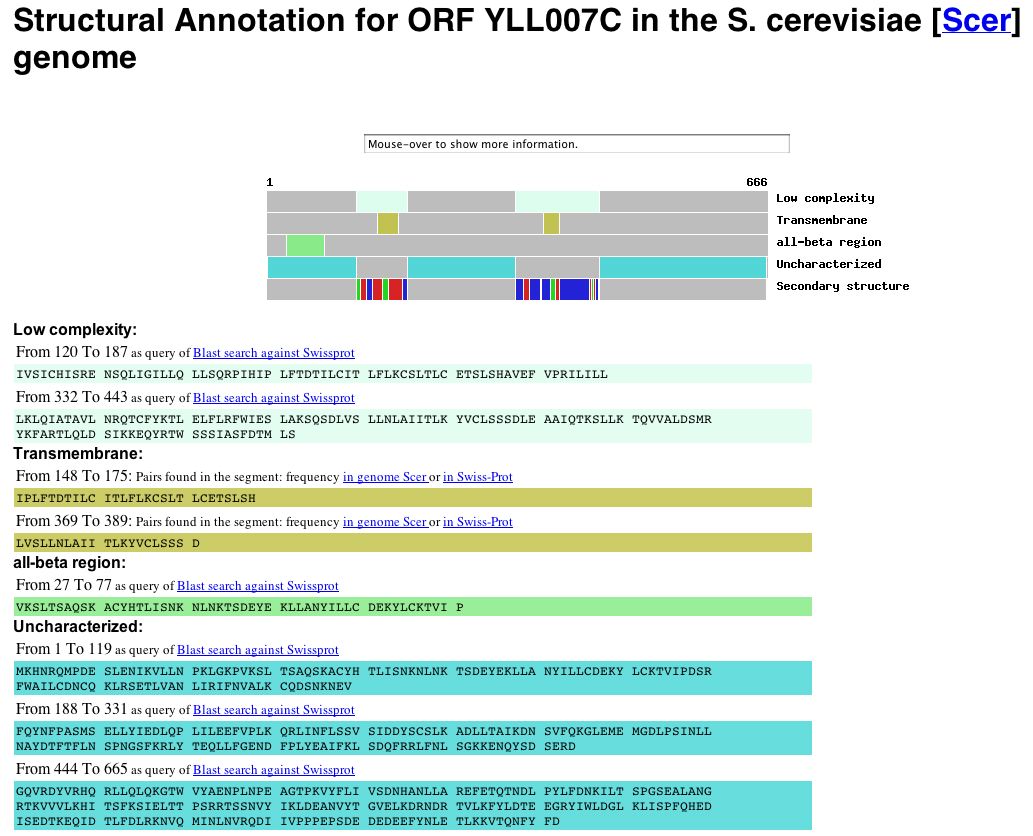 |
| Figure 8: Snapshot of image deicting the structural annotation of the YLL007C gene obtained from Yale Info on the Microarray Global Viewer database. The YLL007C protein is predicted to have two trans-membrane domains (amino acids 148-175 and amino acids 369-389) as well as an all-beta region (amino acids 22-77). The data indicate, however, that the majority of the protein is uncharacterized. |
Information from PROWL: The PROWL database indicated that the YLL007C protein has a molecular mass of 76573.721 Da and an isoelectric point of 5.6. |
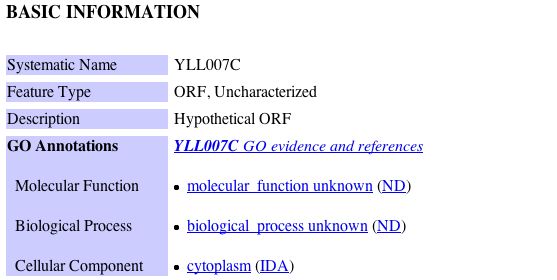
Information from MIPS: The MIPS database indicated that YLL007C is a known protein of unknown function that is localized to the cytoplasm. The database also indicated that YLL007C has genetic interactions with three other proteins. The results of this search can be seen below: |
| Figure 9: Snapshot of image obtained from MIPS database using a search for YLL007C. The database indicated that YLL007C has genetic interactions with yeast proteins GIM4, GIM5 and PAC10. |
In order to determine the function of these proteins that interact with YLL007C, I searched the Saccharomyces Genome Database. The results of this search can be seen below: |
|
| Figure 10: Snapshots of images for information regarding GIM4, GIM5 and PAC10 obtained from the Saccharomyces Genome Database. |
It is important to note that all interactions involving YLL007C that were identified by the MIPS database are genetic interactions, indicating that YLL007C does not physically interact with any of these proteins listed above. Rather, genetic interactions tend to be determined from mutant phenotype data in which redundancy in the genome can be identified. A systemic deletion of YLL007C results in a growth defect phenotype. Yeast that have null mutations of YLL007C accompanied by null mutations of GIM4, GIM5, and PAC10 are all viable but exhibit a slow growth phenotype (Tong et al. 2004). Given this information, it is important to note that all of the proteins with which YLL007C has a genetic interaction are asociated with tubulin binding and tubulin folding. Tubulin is the protein that composes microtubules within the cell. Microtubules determine the position of various membrane enclosed organelles and often direct intracellular transport. Microtubules are also responsible for the formation of the bipolar mitotic spindle during cell division. Proteins that bind microtubules are collectively known as microtubule associated proteins (MAPS). They can act to stabalize of destabilize microtubule elongation in the cell (Alberts et al. 2002). Since microtubules are essential for mitosis in all eukaryotic cells, it is not surprising that yeast mutants involving proteins that are associated with tubulin binding and tubulin folding have a slow growth phenotype. In the previous example with Cox17, those proteins with which it had a genetic interaction were more closely associated with its function than those proteins with which there was a physical interaction. Using this logic, I hypothesize that there is some overlap of functional capacity between YLL007C, GIM4, GIM5 and PAC10. Hence, it is possible that YLL007C is also associated with tubulin binding and tubulin folding in yeast and possibly microtubule assembly. |
Information from DIP: Unfortunately, a search of the Database of Interacting Proteins (DIP) yielded no results for the YLL007C ORF. This indicates that no physical interactions between YLL007C and other yeast proteins have been identified. This corresponds with the data obtained from MIPS, which indicated that only genetic interactions between YLL007C and other yeast proteins have been identified. |
Information from SGD: With no information about YLL007C from DIP, I decided to do a search of the protein-protein interaction information for YLL007C from the Saccharomyces Genome Database. The results of the yeast two hybrid interaction data from the Yeast Resource Center can be seen below: |
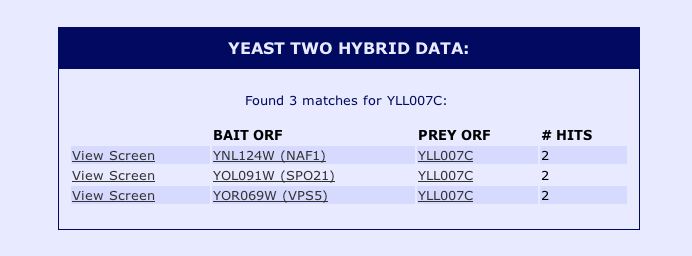 |
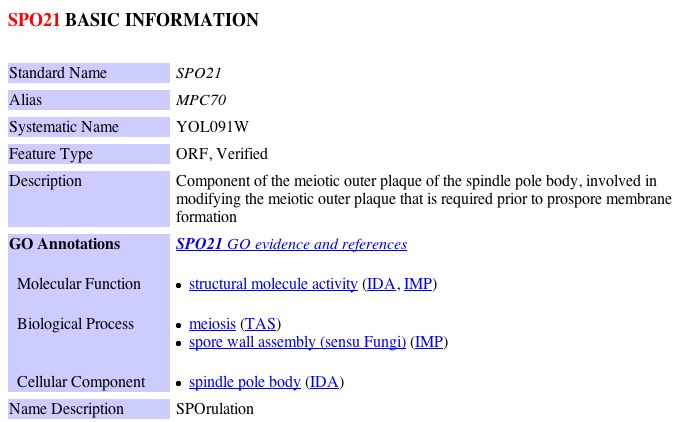
Figure 11: Snapshot image of the yeast two hybrid data for yeast ORF YLL007C obtained from the Saccharomyces Genome Database protein information. These results indicate that the non-annotated YLL007C protein physically interacts with NAF1, SPO21 and VPS5. In order to obtain more information about these proteins implicated in the Yeast Two Hybrid study, I performed a search using the Saccharomyces Genome Database for these proteins. The results can be seen below: |
|
| Figure 12: Snapshot images of information regarding NAF1, SPO21 and VSP5 obtained from the Saccharomyces Genome Database. |
Keeping in mind YLL007C's likely involvement with tubulin binding/folding and my previous predictions regarding the function of YLL007C, I decided to examine possible ways in which each of these proteins could have a physical interaction with YLL007C. NAF1 is a nuclear assembly factor, thus my earlier hypothesis that YLL007C is a nuclear membrane protein could be feasible. Similarly, SPO21 is implicated in meiosis, thus, it would not be unusual for a nuclear membrane protein to have associations with the meiotic outer plaque of the spindle pole body. Despite this limited evidence which does not contradict my previous conclusion, it is difficult to draw a connection between a nuclear membrane protein and VPS5, a protein that is involved in protein transport between the endosome and the Golgi. Thus, in light of this new evidence, I must redefine my previous predictions regarding the cellular function of YLL007C. After investigating the limited data regarding YLL007C further, I came to the conclusion that are there are two conflicting interpretations among the databases regarding its cellular localization. For example, YLL007C is predicted to have at least two transmembrane domains and yet the Saccharomyces Genome Database as well as the MIPS database indicate that YLL007C is localized to the cytoplasm. The information from these databases can be seen below: SGD Informtaion regarding YLL007C:
MIPS Informtaion regarding YLL007C:
Furthermore, other databases which are accessable via the Yeast Microarray Global Viewer (Yale Info) indicate that YLL007C is localized to the nucleus: YALE INFO informtaion regarding YLL007C:
In light of this conflicting information regarding the localization of of YLL007C, I have changed my original prediction regarding the cellular role of this non-annotated ORF. Given the likely association of YLL007C with tubulin, and hence microtubles, it is likely that this protein is not restricted to the nuclear membrane. In addition, the fact that NSF1 is involved in nuclear assembly and SPO1 is implicated in spore wall assembly is consistent with the prediction that YLL007C is involved in microtubule assembly in some capacity, as microtubules are an integral part of the nuclear membrane and any form of nuclear or cell division. |
Information from TRIPLES database: In order to investigate further whether or not my new prediction is valid, I decided to look at the TRIPLES database, otherwise known as the database of TRansposon Insertion Phenotypes, Localization and Expression in Saccharomyces. This database displays data from different transposon insertion events in the yeast genome. When a transposon-encoded lacZ (a reporter geneP) is inserted into a particular yeast ORF, b-gal filter assays can be used to determine when a given gene is expressed during the yeast life cycle. A search for the YLL007C ORF indicated that an insertion event did occur at this locus in one the experiments. The results of this experiement can be seen below: |
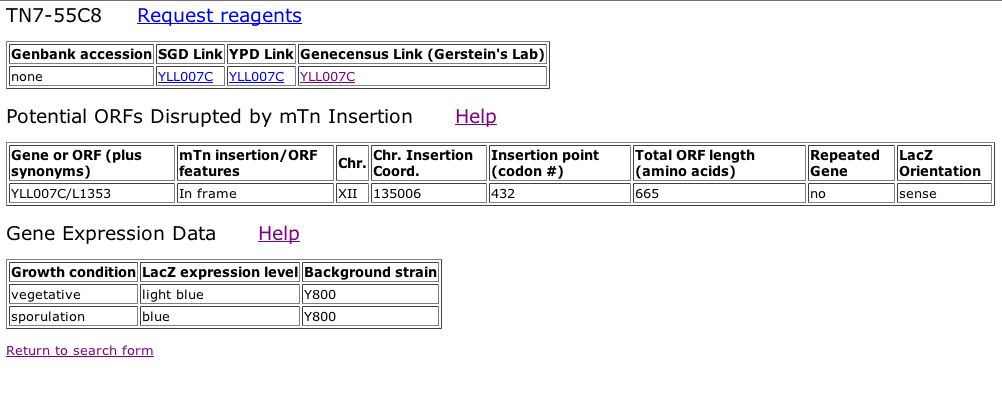 |
| Figure 13: Snapshot image obtained from TRIPLES database. This image displays the ORF disrupted by the mTn insertion and the Gene Expression Data in terms of LacZ expression levels during different growth conditions of the cell cycle. The LacZ expression levels are determined from the observed intensity of blue staining after growth on medium containing X-gal. Stain intensity is scored as either faint blue stain, light blue color, blue or intense blue staining. Given this scale it is clear that expression of the YLL007C ORF is induced during sporulation. |
| Although this information from TRIPLES did not provide any conclusive information, induction of YLL007C during sporulation is not inconsistent with my prediction that the YLL007C protein is involved in tubulin binding and folding. Microtubule assembly is an important aspect of spore formation in yeast, thus, if YLL007C were to be associated with tubulin it is likely that it would be induced during spore formation. |
Informtaion from PDF Files: No information regarding YLL007C could be found in the PDF files (Aging, Benno Figure 1, Benno Figure 2, Degredation) that were dicussed in class. |
Future Experiments:
There are several experiments involving YLL007C that should be performed. The first thing that should be resolved is the conflicting information regarding the cellular localization of this protein. In order to get a more accurate picture of YLL007C's localization within the cell, I would start by generating a monoclonal antibody to YLL007C to use in immunoflourescence studies. A secondary antibody with a flourescent marker (which would bind this monoclonal antibdody) would also be needed. Using immunoflouresence, a more accuarte picture of YLL007C's location in the cell might be able to be determined. In addition, this experiment would also allow one to get a better idea of whether or not this protein is indeed a transmembrane protein. If flourescene was only visible around the periphery of the nucleus, one could determine that YLL007C is a nuclear membrane protein. The expression data seen in the previous assignment indicated that YLL007C was induced and repressed in various stages of the yeast cell cycle. Although mRNA expression levels can provide insight into when proteins are present in high amounts or low amounts, this data does not give any precise quantification. One way to better determine the quantity of protein present at a given time would be to use the ICAT method. Using two yeast populations that were at different stages of the cell cycle, one could label YLL007C with the heavy ICAT reagent in one population and the light ICAT reagent in another population. It would also be useful to have the ICAT data regarding the proteins with which YLL007C physically and genetically interacts from these same two populations of cells. With this information, the amount of protein present in the two given conditions could be quantified, and YLL007C could be more conclusively associated with various cellular processes. In another effort to determine which future experiements should be done to further characterize the cellular role of YLL007C, I did some further investigation on the proteins that were determined to have a genetic interactions with YLL007C. A study conducted by Geissler et al. in 1998 found that null mutants of the GIM family of proteins (including GIM4 and GIM5) produced a synthetially lethal phenotype with Tub4-1, which encoded a mutated yeast gamma-tubulin. Based upon this study, it was surmised that the Gim proteins form a protein complex that promotes formation of functional alpha- and gamma-tubulin. In order to determine whether or not YLL007C plays a role similar to those proteins in the GIM family, I would conduct an experiment to determine whether or not a null YLL007C mutant also produced a synthetically lethal phenotype with Tub4-1. If the study yielded the same result, one could conlcude that YLL007C is likely to be a part of the protein complex that forms functional alpha and gamma tubulin.
|
References: [DIP] Database of Interacting Proteins. 2004 Nov 15. http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3 Accessed 2004 Nov 17. [KEGG] Kyoto Encyclopedia of Genes and Genomes http://www.genome.jp/kegg/genes.html Accessed 2004 Nov. 17. [MIPS] Munich Information Center for Protein Sequences. 2004 Nov 15. http://mips.gsf.de/genre/proj/yeast/index.jsp. Accessed 2004 Nov 17. [PDB] Protein Data Bank. 1U96. http://www.rcsb.org/pdb/cgi/explore.cgi?pid=222991100880390&page=0&pdbId=1U96 Accessed 2004 Nov 17. [PROWL] Protein Info. http://129.85.19.192/prowl/proteininfo.html Accessed 2004 Nov 17. [SGD] Saccharomyces Genome Database. 2004 Nov 15. SGS1/YMR190C. http://db.yeastgenome.org. Accessed 2004 Nov 15. [TRIPLES] TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces. http://ygac.med.yale.edu/triples/basic_search.asp. Accessed 2004 Nov 17. [yMGV] Yeast Microarray Global Viewer. http://www.transcriptome.ens.fr/ymgv/ Accessed 2004 Nov. 17. Alberts B, Alexander J, Lewis J, Raff M, Roberts K, Walter P. 2002. The Cell: New York. p. 907-982. Beers J, et al. (1997) Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J Biol Chem 272(52):33191-6 Campbell AM, Heyer LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings: San Francisco. p.162-204. Geissler S, et al. (1998) A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17(4):952-66
|