This web page was produced as an assignment for an undergraduate course at Davidson College.
SGS1 and YMR187C: Proteomic Data and Interpretation
Overview:
A proteome, or the complete set of proteins in a cell, tissue, or organism at any particular point (Campbell and Heyer 2003), can provide much insight about the biological importance of particular proteins. The genes SGS1 and YMR187C are both located on Saccharomyces cerevisiae (yeast) chromosome XIII. In previous assignments, I have explored the sequences and expression patterns of SGS1 and YMR187C. SGS1 is involved in ATP-dependent DNA helicase activity, meaning that it aids in unwinding the double helix so that processes like DNA replication and mRNA synthesis can occur. SGS1 also plays a role in maintaining genomic stability, chromosome organization and biogenesis, meiotic chromosome segregation, and mitotic sister chromatid segregation. Researchers predict that a mutation in SGS1 might lead to rapid aging in yeast, which could perhaps lead to further our knowledge of longevity and aging.
YMR187C is a non-annotated gene, so its molecular function, biological process, and cellular component are unknown. From sequence analysis, I predicted that YMR187C is a transmembrane protein and either could be a transcription factor affecting SGS1, could act similarly to the monkey histocompatibility complex, or could play some role in important DNA processes like mitosis, meiosis, replication, and/or transcription. From gene expression data, I gathered that YMR187C probably plays more than one role in the life of yeast. It could be a regulator of stress responses, a transcription factor affecting DNA
polymerase II, a protein involved in cytokinesis and protein organization, or could be involved in important DNA processes. These hypotheses will be re-examined through the use of proteomic data.
| Note: My comments and conclusions will be enclosed in gray boxes like this one, so that readers can easily distinguish my thoughts from the purely factual information. |
SGS1 Proteome:
Protein Structure and Physical Properties
Only a portion of the SGS1 protein has been crystallized, so a partial representation of its structure has been determined. Figure 1 below is a chime image of the HDRC domain of SGS1, which ranges from amino acids 1271 to 1351 of its sequence. The helicase and RNaseD C-terminal (HRDC) domain is an 80-amino acid domain
usually found at the C-terminus of RecQ helicases and RNase D homologs from
various organisms, including human, yeast and bacteria (ExPASy 2004). The HRDC domain is
involved in the binding of DNA to specific DNA structures (e.g. long-forked
duplexes and Holliday junctions) that are formed during replication,
recombination or transcription (Swiss Model Repository of ExPASy 2004). Figure 2 is a visual representation of the entire amino acid sequence of the protein and the portion of that sequence represented by the model in Figure 1.
| |
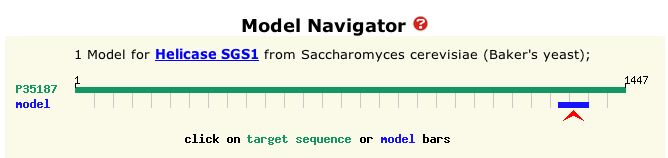 |
Figure 1: Chime image of known region of SGS1 protein. This protein is a total of 1447 amino acids, but the sequence of the structure above correlates to amino acid positions 1271 to 1351 of the protein's sequence. (PDB 2004)
(Can't see the image? You probably need a Chime plug-in.) |
Figure 2: Graphical depiction of amino acid sequence of SGS1 protein and crystallized portion. The blue bar represents the sequence of the model protein shown in Figure 1. Total length is 1447 amino acids, and the blue bar demarcates positions 1271 to 1351 of the total protein sequence. (ExPASy 2004)
|
| From Figure 2, it is evident that the protein in Figure 1 is a very small portion of the entire sequence. It is probably an important region, however, because according to ExPASy (2004), the HDRC domain is conserved in many other helicases. |
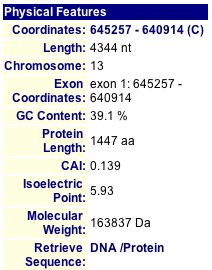
MIPS (2004) and SGD (2004) both provide a quick view of pertinent information about SGS1 (see Figures 3 and 4).
 |
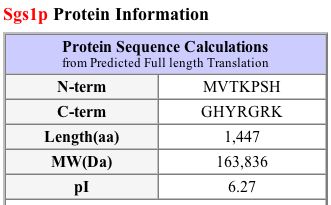 |
Figure 3: Physical Features of SGS1. The protein length is 1447 amino acids, isoelectric point is 5.93, and molecular weight is 163837 Daltons. (MIPS 2004) |
Figure 4: Protein information about SGS1. The protein length is 1447 amino acids, isoelectric point is 6.27, and molecular weight is 163836 Daltons. (SGD 2004) |
I also used PROWL (2004) to run a sequence analysis of SGS1. It determined the molecular mass to be 163735.109 Daltons and the isoelectric point to be 5.9.
| Figures 3 and 4, along with the PROWL data, show that proteomic information is not always consistent across different databases. Although the lengths are the same and the molecular weights are fairly similar, the isoelectric point listed on SGD seems rather different than the ones that I found at MIPS and PROWL (2004). |
Protein Function
The TRIPLES database provides insight to a protein's function through the use of experiments in which transposable elements (mTn's) are used to disrupt genes. See TRIPLES information for SGS1 in Figure 5.
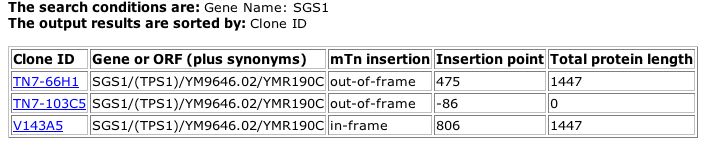 |
| Figure 5: TRIPLES (2004) output for SGS1. The second entry shows that a transposable element was insterted at point -86, which lies in the 5' region (the promoter), and as a result, no protein was produced (length=0). The other two experiments did not alter the normal length of the protein. |
| In Figure 5, the first and third interruption experiments did not alter the length of the SGS1 protein, so they do not provide much helpful information. In the second experiment, however, the transposable element was inserted at bp 645343. SGS1 spans from base pairs 630914 to 655257. Since DNA is transcribed in the 5' to 3' direction, the 5' region is likely the promoter area. The insertion point is also labeled as codon number -86. In general, the first codon that codes for the protein is labeled number one, while codons in the promoter are labeled with negative numbers. The interruption of the sequence at codon -86 indicates that this portion of the promoter sequence is required for proper protein production. It is obvious that the interruption of a promoter will result in a the halt of transcription. Thus, the TRIPLES data provides little additional insight into the function of the SGS1 protein. |
With a basic Google search, I found the Saccharomyces cerevisiae Genomic Phenotyping Database which contained information about the viability of SGS1 in a knockout experiment. This group of researchers used yeast deletion strains and tested them against the following macromolecular damaging agents: methyl methanesulfonate (MMS), tert-butyl hydroperoxide (t-BuOOH), 4-nitroquinoline-N-oxide (4NQO) and ultraviolet radiation (UV). Strains were grown in 96-well plates, and images were taken after 60 hours of growth. They compared the growth of the knockout to the growth of the wild type for each treated plate, and also examined the growth of the knockout on untreated media to make sure that it was not totally inviable. See Figure 6 below for experimental data.
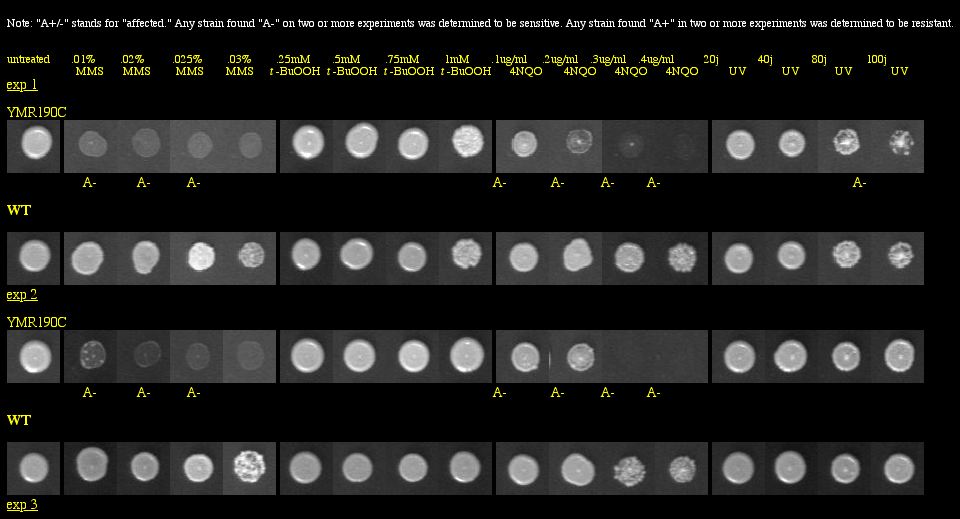 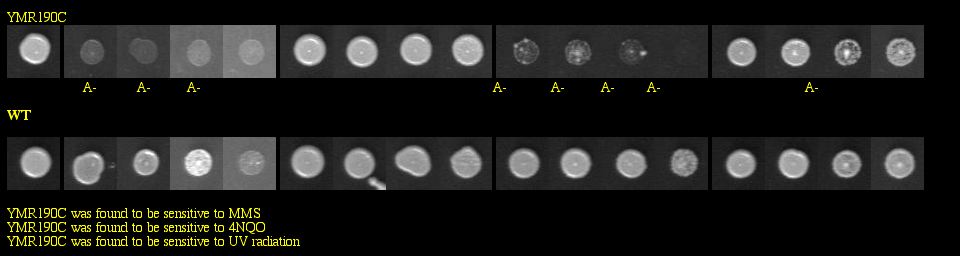
|
Figure 6: Phenotype macroarray data for SGS1 deletion strain, taken from Saccharomyces cerevisiae Genomic Phenotyping Database (2004). "A+/-" stands for affected, and any strain with A- on two or more experiments was determined to be sensitive. YMR190C (or SGS1) was found to be sensitive to MMS, 4NQO, and UV radiation. Note: The resolution of your computer may need to be adjusted to visualize the scoring properly. Visit the database for instructions about monitor resolution.
|
| Figure 6 shows that the SGS1 deletion strain is vulnerable to three macromolecular damaging agents. These data strongly confirm SGS1's role in maintaining genomic stability, particularly because the untreated deletion strains were viable. |
Protein-Protein Interactions
In the Database of Interacting Proteins (DIP), I found a map of SGS1's protein interactions. Here, an "interaction" is defined by the experimental identification of two amino acid chains binding to each other (DIP 2004). Figure 7 below depicts the protein interaction map for SGS1, and the key for interpretation is shown in Figure 8.
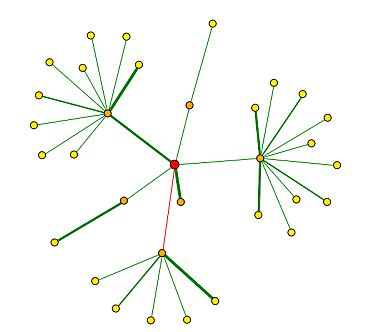 |
 |
Figure 7 : Map of SGS1 protein interactions. SGS1 is represented by the red circle, or central node. (DIP 2004) |
Figure 8 : Key for interpretation of DIP maps. (DIP 2004) |
I created Table 1 below in order to show the genes represented by the first shell nodes (orange circles in Figure 7) and their functions.
| Protein name |
Function (found on DIP 2004) |
| Top2p |
DNA topoisomerase (ATP-hydrolyzing) |
| Swd3p |
probable GTP-binding protein
|
| Top3p |
DNA topoisomerase III |
| Msh6p |
DNA repair protein YDR097c |
| Mlh3p |
hypothetical protein YPL164c |
| Rad16p |
DNA repair protein |
| Table 1: Proteins of first shell nodes in Figure 7. Starting from the 12:00 position (with SGS1, or the red circle as the center point), the proteins are listed in a clockwise fashion. |
| Using Figure 8 to interpret Figure 7, it seems that SGS1 has strongly verified core interactions with both Rad16p and Top3p. SGD (2004) says that Rad16 is involved in DNA-dependent ATPase activity, recognizes DNA damage, and performs nucleotide excision repair. SGD reports that Top3 is involved in DNA topoisomerase type I activity, and is involved in meiotic recombination, regulation of DNA recombination, and telomerase-dependent telomere maintenance. Using guilt by association (meaning that proteins that physically interact with each other act in similar manners), I think that these data seem consistent with the previously determined functions of SGS1: maintaining genomic stability, chromosome organization and biogenesis, meiotic chromosome segregation, and mitotic sister chromatid segregation. |
MIPS (2004) also presents a summary of SGS1's physical and genetic interactions, which can be seen in Figure 9.
 |
Figure 9 : Listing of physical and genetic interactions of SGS1, according to MIPS 2004. |
| It seems as though there are some slight discrepancies between the information in Figures 7 and 9. Not all of the proteins found in Figure 7 are listed in Figure 9, and vice versa. Top3 still retains a strong presence, however, so I am fairly convinced that it interacts with SGS1. I was surprised, though, that Top2 was found in Figure 7 but not in Figure 9, and that Top1 was found in Figure 9 but not in Figure 7. (Note: The "Visualize using GenRE-Int" link was broken, so I was unable to find more information about the high-throughput data.) |
Upon further inspection of SGS1's physical interactions, I found relevant yeast two-hybrid (Y2H) data on SGD (2004). Yeast two-hybrid is a method in which a particular protein is used as a "bait" to see which other "prey" proteins interact with it. See Figure 10 below for results.
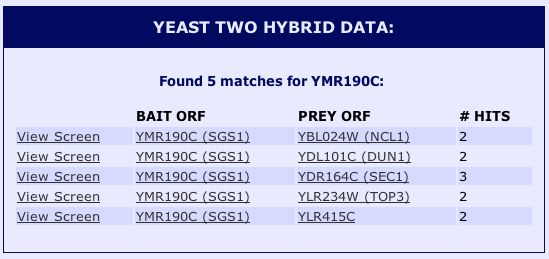 |
Figure 10 : Yeast two-hybrid data for SGS1. (SGD 2004) |
| The Y2H data in Figure 10 present proteins not mentioned by MIPS or DIP. According to SGD (2004), NCL1 is a nuclear protein, DUN1 is a cell-cycle checkpoint serine-threonine kinase required for DNA damage-induced
transcription of certain target genes, and SEC1 is a sm-like protein involved in docking and fusion of exocytic vesicles through
binding to assembled SNARE complexes at the membrane. YLR415C is a non-annotated gene. Although these interactions were not discovered by the other sites, they seem to follow along the lines of SGS1's interactions with proteins required for important DNA processes like replication, transcription, and translation. |
Ultimately, the proteomic data seem to confirm SGS1's predetermined molecular functions and biological processes. Nevertheless, I think it would be beneficial to determine the entire structure of the protein. To achieve this, researchers should attempt to discover other binding sites or conserved domains within the sequence. They could then use the protein sequence to make structural predictions and conceptualize the entire protein.
It might also be interesting to use a fluorescent marker protein to visualize SGS1 during it's roles in mitosis and meiosis. Scientists could bind a specific primary antibody to the protein sequence somewhere, then expose dividing cells to an appropriate secondary antibody marked with a fluorescent protein. This way, the protein could be better localized and visualized in its biological processes. I predict that the images would show SGS1 functioning during meiotic chromosome segregation and mitotic sister chromatid segregation.
|
It should be noted that found no pertinent information about the SGS1 protein at the following sites:
Pathcalling Yeast Interactive Database
Enzymes and Metabolic Pathways
Swiss 2D Page Database
German Protein Structure Factory
GenomeNet
PUMA2
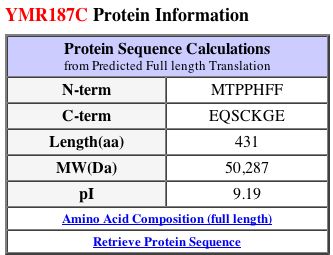
YMR187C Proteome
Protein Structure and Physical Properties
Although it is suspected that YMR187C is a transmembrane protein, its 3D structure has not yet been determined. I found no data for this ORF on PDB (2004).
MIPS and SGD have short charts of information about YMR187C, shown below in Figures 11 and 12.
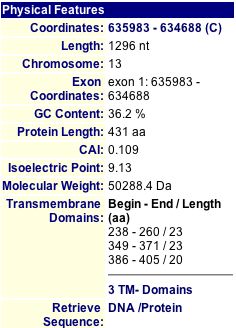
|
|
Figure 11: MIPS information about the YMR187C protein (2004). |
Figure 12: Protein information about YMR187C from SGD (2004). |
By running a sequence analysis on PROWL (2004), I discovered that the molecular mass of the protein is 50254.938 Daltons and the isoelectric point is 9.6, much higher than that of SGS1.
Just like SGS1, the isoelectric point data differ from each other. MIPS and SGD seem to report values that are similar, but PROWL gives a greater value. The molecular weights are also different across the three databases. I think it is interesting to note that the SGS1 and YMR187C proteins are very different from each other in many aspects: length (1447 and 431 amino acids each, respectively), isoelectric point (around 6 and 9, respectively), and molecular weight (163,000 Daltons and 50,000 Daltons, respectively). Clearly, they must have different functions, yet they could still be related in some way.
|
Protein Function
The TRIPLES database showed just one experiment for YMR187C, shown in Figure 13.
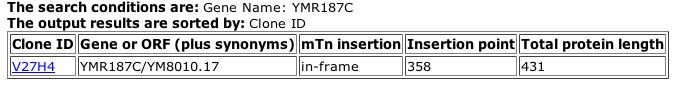 |
| Figure 13: TRIPLES data for YMR187C. The transposable element did not cause a change in the normal protein length (2004). |
| Although transposable elements can often be used to help determine important portions of a protein or sequence, the results above provide little insight to the role of YMR187C in the life of yeast because the length of the protein was not altered by the mTn. |
I found a YMR187C knockout experiment on the Saccharomyces cerevisiae Genomic Phenotyping Database. See Figure 14 below for results.
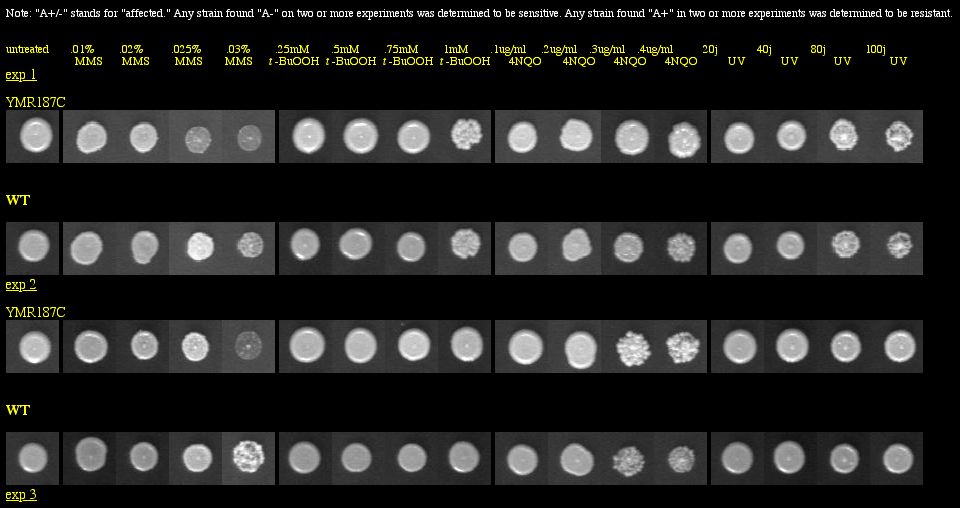 
|
| Figure 14: Phenotype macroarray data for YMR187C deletion strain, taken from Saccharomyces cerevisiae Genomic Phenotyping Database (2004). "A+/-" stands for affected, and any strain with A- on two or more experiments was determined to be sensitive. The YMR187C knockout was not found to be responsive to any treatment. Note: The resolution of your computer may need to be adjusted to visualize the scoring properly. Visit the database for instructions about monitor resolution. |
| The phenotype macroarray data of the YMR187C deletion strain show that the YMR187C protein is not required for viability when the strains are exposed to macromolecular damaging agents. Thus, YMR187C cannot be vital to important DNA processes, because if it were, we would expect to see sensitivity in the experiment above, similar to what was seen with SGS1 in Figure 6. |
Protein-Protein Interactions
DIP (2004) provides a physical interaction map of proteins that bind with YMR187C. See Figure 15 below for map, Figure 8 above provides the key for interpretation.
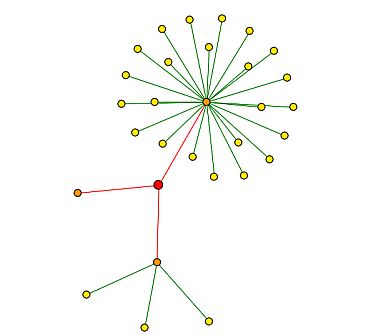 |
Protein name |
Function and location (found on SGD 2004) |
Jsn1p |
member of Puf family of RNA-binding proteins; interacts with mRNAs encoding membrane-associated proteins (Chr X) |
Lys14p |
transcription activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer (Chr IV) |
Rsa3p |
likely role in ribosomal maturation; required for accumulation of wildtype levels of large (60s) ribosomal subunits; binds to helicase Dbp6p in pre-60s ribosomal particles in nucleolus (Chr XII) |
|
| Figure 15: DIP (2004) interaction map of YMR187C. YMR187C is depicted by the red circle, or central node. All three nodes are indicated with red edges, meaning that they are unverified high-throughput interactions (2004) |
Table 2: Proteins of first shell nodes (orange circles) in Figure 15. Starting from the 12:00 position (with YMR187C as the center point), the proteins are listed in a clockwise fashion. |
| Figure 15 leads me to believe that YMR187C could be a an RNA-binding protein, a transcription activator, or could have an effect on ribosomes. Nevertheless, the interactions were discovered through high-throughput methods only, and are not verified by other experimental evidence. |
I found a list of the physical interactions of YMR187C on MIPS (2004):
 |
| Figure 16: Physical interactions listed for YMR187C on MIPS (2004). |
| Figure 16 shows the exact same proteins shown in Figure 14, just in a different fashion. For me, the visual map of interactions in Figure 14 is more appealing. Still, it does not provide much new information in my search for YMR187C's true function. |
Finally, I looked for yeast two-hybrid data for YMR187C on SGD (2004) with no success. The YRC (Yeast Resource Center) and additional results (2004) databases did not list any experiments involving YMR187C, either.
After much thought, have modified my hypotheses about YM187C's functions. I doubt that it acts as a transcription factor, because it contains a number of transmembrane domains. It seems to me that transcription factors are usually proteins that can be found floating around in solution, not adhered to membranes. YMR187C could still play a role in important DNA processes like mitosis, meiosis, replication, and/or transcription, however, by being involved in cytokinesis and protein organization. I doubt that it is crucial as a regulator of stress responses, because the deletion strain was viable in the phenotype macroarray experiment shown in Figure 14. By process of elimination and the acquisition of new information, I propose that YMR187C's most important roles are in cytokinesis and protein organization.
Future studies ought to explore more about the 3D structure of the protein to better characterize its biological processes. Sequence analysis shows the presence of a zinc finger domain, so perhaps scientists could determine the structure of that portion of the protein, at least, through the study of other zinc finger domains.
More phenotype macroarray experiments would also be helpful. Maybe the YMR187C deletion strain is viable on the media tested above in Figure 14, but perhaps it is inviable under many other conditions. If it is inviable under other conditions, perhaps those particular conditions could be tested with RNAi experiments to determine the exact point in time when the protein is vital to the life of the strain. I predict that the YMR187C deletion strain must be inviable under some condition, maybe one that affects the cell particularly during division.
Along the same lines, additional TRIPLES experiments would be beneficial. Only one mTn was used for YMR187C, so maybe more experiements might yield more meaningful results. I think there must be some region of the sequence that is vital to the production of the full-length protein, perhaps the zinc finger domain.
One last option for study is to individually confirm the physical interactions presented in Figures 15 and 16. High-throughput methods are excellent for surveying an entire genome, or a large amount of information. They fail, however, when specificity is needed. Maybe the YMR187C protein and another suspected to interact with it physically could each be marked with different colored, specific, fluorescently labeled antibodies. The results would show one color or two, depending upon the interaction of the proteins. I presume that the experiment would confirm the physical interactions previously determined by high-throughput methods, and might lead to further discovery physical interactions with YMR187C if additional proteins were also tested.
Now that high-throughput methods have scratched the surface of the genomic era, it's time to start digging deeper.
|
It should be noted that found no pertinent information about the YMR187C protein at the following sites:
Pathcalling Yeast Interactive Database
Enzymes and Metabolic Pathways
Swiss 2D Page Database
German Protein Structure Factory
GenomeNet
PUMA2
References:
[ExPASy] Expert Protein Analysis System. 2004 Nov 15. http://www.expasy.org/. Accessed 2004 Nov 15.
[DIP] Database of Interacting Proteins. 2004 Nov 15. http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3 Accessed 2004 Nov 15.
[MIPS] Munich Information Center for Protein Sequences. 2004 Nov 15. http://mips.gsf.de/genre/proj/yeast/index.jsp. Accessed 2004 Nov 15.
[PDB] Protein Data Bank. 1D8B. http://www.rcsb.org/pdb/cgi/explore.cgi?pid=203151097204076&pdbId=1D8B. Accessed 2004 Nov 15.
[PROWL] Protein Info. http://129.85.19.192/prowl/proteininfo.html Accessed 2004 Nov 16.
[SGD] Saccharomyces Genome Database. 2004 Nov 15. SGS1/YMR190C. http://db.yeastgenome.org. Accessed 2004 Nov 15.
[TRIPLES] TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces. http://ygac.med.yale.edu/triples/basic_search.asp. Accessed 2004 Nov 16.
Campbell AM, Heyer LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings: San Francisco. p.107-109.
Saccharomyces cerevisiae Genomic Phenotyping Database. YMR187C. http://genomicphenotyping.mit.edu/pages/source2.html. Accessed 2004 Nov 17.
___________________________________________________________________________________________________________________________________________________
For more information, please refer to the Genomics Home Page or to the Davidson College Home Page.
Questions or comments? E-mail me: "jehoekstra" at "davidson.edu".
Back to my genomics home page.
Page last updated 2004 Nov 19 by J. Hoekstra.