*This web page was produced as an assignment for an undergraduate course at Davidson College*
My Two Favorite Saccharomyces cerevisiae (Yeast) Genes
Annotated Gene |
Non-annotated Gene |
Vas 1 |
YGR093W |
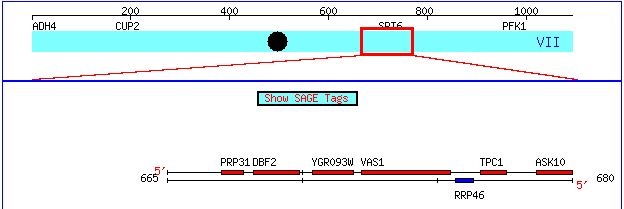
Figure 1. This image, taken from http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?chr=7&beg=667000&end=767000, zooms in on the chromosomal location of the gene Vas1 and its non-annotated neighbor YGR093W.
Introduction
In the above snapshot, the coding region of Yeast Chromosome VII that contains my two favorite yeast genes. Vas 1 is an annotated gene, meaning the protein that it encodes has been well characterized. The neighboring ORF, YGR093W, is a non-annotated gene. That is, while YGR093W appears to be a coding sequence for some protein, the structure and function of this protein are currently hypothetical. On this webpage, I will present all pertinent information about the function of the Vas 1 gene and provide educated predictions about the predicted product of theYGR093W ORF.
Annotated Gene: Valyl-tRNA Synthetase (Vas1)
Chromosomal Location
Vas1 is located on Chromosome VII of the Yeast genome from bp 672190 - bp 675504. This top strand of DNA is the coding strand for this gene (Figure 1).
Biological Process
Aminoacyl tRNA synthesases are a varied family of enzymes that all perform the same general function. These enzymes catalyze the joining of tRNA molecules to the appropriate amino acid, a key prerequisite step for proper protein translation (Figure 2). Each enzyme is specific to a particular amino acid (wikipedia 2005).

Figure 2. Taken from http://en.wikipedia.org/wiki/Aminoacyl_tRNA_synthetase, this snapshot shows the general two-step reaction catalyzed by aminoacyl tRNA synthetases.
Vas1 encodes the Valyl tRNA Synthetase, which catalyzes the formation of valyl-tRNA.
Molecular Function
The Vas1 protein shares a similar molecular function with other aminoacyl tRNA Synthetases (Figure 3).
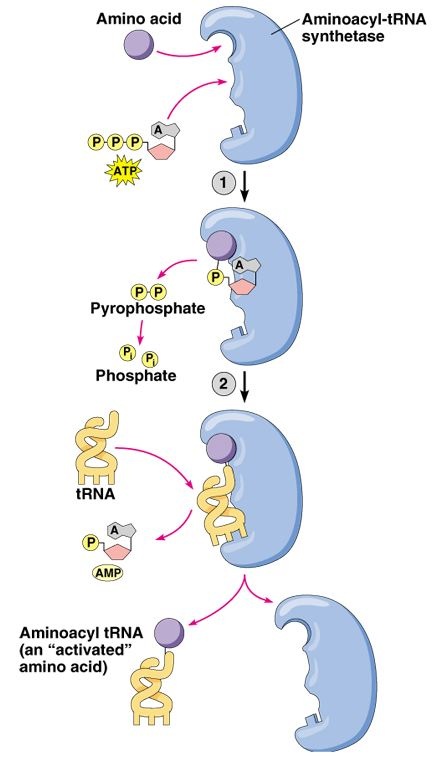
Figure 3. Permission pending for this figure from Addison Wesley Longman. This figure depicts the general molecular function of an aminoacyl tRNA synthetase. First, the enzyme binds the amino acid and joins it to a molecular of AMP while cleaving two phosphate groups from a molecule of ATP. Next, the enzyme binds the aminoacyl portion of this complex to an appropriate tRNA molecule while releasing the AMP molecule.
First, the Vas1 protein forms a valyladenylate complex. Then, the enzyme transfers the valyl-portion of the complex to the appropriate tRNA molecule. However, Vas1 does not perfectly discriminate between valyl and threonyl, leading to the formation of an unusable threonyladenylate complex (Baldwin and Berg, 1966).
Cellular Component
The protein product of this gene is found in both the cytoplasm and the mitochondria of Saccharomyces cerevisiae. This gene is alternatively spliced, forming two mRNA transcripts of different lengths. The longer transcript, which is thought to be the mitochondrial version, begins translation with a methionine at position 1. The shorter, cytoplasmic transcript begins with methionine at amino acid position 47 (of the longer transcript) (Chatton et al., 1987).
Nucleotide Sequence |
Amino Acid Sequence |
| ATGAATAAGTGGTTAAACACATTATCTAAGACATTCACTTTTCGGCTTTTGAACTGTCAT TATAGGCGATCATTACCACTTTGTCAAAACTTTTCTCTGAAGAAGTCGTTAACTCATAAT CAAGTCAGGTTCTTTAAAATGAGCGATCTTGATAATTTGCCTCCAGTTGACCCAAAGACT GGTGAGGTCATCATTAATCCGTTAAAGGAAGATGGCTCTCCAAAGACTCCTAAGGAAATT GAAAAAGAGAAGAAAAAGGCTGAAAAACTGTTAAAGTTCGCTGCCAAACAAGCTAAAAAA AATGCTGCTGCCACCACAGGTGCATCTCAAAAGAAACCTAAGAAAAAGAAGGAAGTTGAG CCAATCCCTGAATTTATTGACAAAACTGTTCCAGGTGAGAAAAAAATCTTAGTATCCTTG GATGATCCGGCTTTAAAAGCTTATAACCCTGCTAACGTTGAAAGTTCTTGGTATGACTGG TGGATCAAGACTGGTGTTTTTGAACCTGAGTTTACCGCTGATGGTAAGGTTAAACCAGAA GGTGTATTTTGCATTCCAGCACCTCCACCAAACGTCACTGGTGCCTTACATATTGGTCAT GCTTTGACTATTGCTATCCAAGATTCTTTGATCAGATATAACAGAATGAAAGGTAAAACT GTCTTATTCTTGCCAGGTTTCGACCATGCTGGTATTGCTACTCAGTCCGTTGTGGAGAAG CAAATCTGGGCTAAGGACAGAAAGACTAGACATGACTATGGAAGAGAAGCTTTTGTTGGT AAGGTCTGGGAATGGAAAGAGGAATACCATAGCAGAATTAAGAACCAAATTCAAAAATTG GGGGCTTCTTATGATTGGAGCCGCGAAGCTTTCACTTTGAGTCCAGAATTGACCAAGTCT GTTGAAGAAGCTTTTGTTAGACTACATGATGAAGGTGTTATTTATCGTGCGTCCAGATTA GTTAATTGGTCTGTTAAATTGAATACCGCTATCTCTAATTTGGAAGTCGAAAATAAGGAC GTTAAAAGTAGAACGCTTTTATCAGTCCCAGGCTATGATGAAAAGGTTGAATTTGGTGTT TTAACATCATTTGCTTATCCAGTTATCGGTAGCGATGAAAAACTGATCATTGCTACAACT AGACCTGAAACTATATTTGGTGATACTGCCGTTGCAGTTCATCCTGATGATGACCGTTAC AAACACTTGCATGGTAAGTTCATCCAACATCCTTTCTTACCAAGAAAAATTCCAATTATC ACCGACAAGGAAGCTGTTGACATGGAATTCGGTACTGGTGCCGTTAAGATCACTCCAGCC CATGACCAAAACGATTACAATACCGGTAAGCGTCACAATTTGGAATTCATCAATATTTTG ACTGACGATGGTTTATTAAACGAGGAGTGTGGTCCAGAGTGGCAAGGCATGAAGAGGTTT GATGCCAGAAAGAAGGTCATTGAGCAGCTGAAGGAAAAGAACCTATACGTTGGCCAAGAA GATAATGAAATGACCATTCCAACTTGTTCCAGATCTGGTGACATTATTGAACCTTTATTG AAACCTCAATGGTGGGTTTCTCAAAGTGAAATGGCCAAAGATGCTATTAAGGTTGTTAGG GATGGTCAAATTACCATCACCCCCAAATCTTCTGAGGCTGAATATTTCCATTGGTTGGGT AACATCCAAGATTGGTGTATTTCCAGACAATTATGGTGGGGTCATCGTTGTCCAGTTTAC TTTATTAATATCGAAGGCGAAGAACACGATAGAATTGATGGTGACTATTGGGTTGCTGGT AGGAGCATGGAGGAAGCTGAAAAGAAGGCTGCTGCCAAATACCCTAATTCCAAATTTACT CTGGAACAAGATGAAGATGTTTTAGACACCTGGTTCTCGTCCGGTTTGTGGCCTTTCTCC ACTTTGGGTTGGCCAGAGAAGACTAAAGACATGGAAACTTTTTACCCCTTTTCTATGTTG GAAACTGGTTGGGATATTCTTTTCTTCTGGGTTACTAGAATGATTCTATTGGGCTTAAAA TTGACCGGTTCAGTTCCATTCAAGGAAGTTTTCTGCCACTCTTTAGTCCGTGACGCTCAA GGTCGTAAGATGTCTAAATCTTTAGGTAATGTTATTGACCCACTAGACGTTATTACTGGT ATTAAGTTGGATGATTTGCATGCAAAATTATTACAAGGTAACTTAGATCCAAGAGAAGTT GAAAAAGCTAAGATCGGTCAAAAGGAATCCTACCCTAACGGTATTCCTCAATGTGGTACC GATGCTATGAGGTTTGCATTATGTGCTTATACCACTGGTGGTCGTGATATTAACTTAGAT ATCTTACGTGTCGAAGGTTACAGAAAGTTCTGTAACAAAATCTACCAAGCTACCAAGTTT GCATTGATGAGACTCGGTGACGATTATCAACCACCTGCCACTGAAGGTCTATCAGGTAAC GAATCCTTGGTTGAAAAATGGATCTTGCACAAGCTGACTGAAACCTCGAAAATTGTCAAT GAAGCTCTAGATAAACGTGACTTCTTGACGTCCACTAGCAGTATTTACGAATTCTGGTAT TTGATTTGTGATGTTTACATCGAGAACTCTAAATACTTGATTCAAGAAGGCTCTGCTATT GAAAAGAAGTCCGCAAAGGATACATTGTATATCTTGCTGGACAACGCTTTGAAATTAATC CATCCATTCATGCCATTCATTTCTGAAGAAATGTGGCAAAGACTTCCAAAGCGTTCCACT GAGAAGGCTGCCTCAATTGTAAAAGCTTCTTATCCAGTTTACGTATCTGAGTACGATGAT GTCAAATCGGCCAATGCTTACGACTTGGTCTTGAACATTACCAAAGAAGCTCGTTCCTTG TTATCTGAGTACAATATTTTGAAGAATGGTAAGGTTTTCGTTGAATCTAACCACGAGGAA TACTTCAAAACTGCTGAAGATCAGAAAGATTCTATTGTCTCGTTGATCAAGGCCATCGAC GAAGTCACTGTTGTTCGTGATGCTTCCGAAATTCCAGAAGGTTGCGTATTGCAATCTGTT AACCCAGAAGTCAATGTACATCTTCTCGTCAAGGGACACGTTGATATTGATGCTGAAATT GCGAAAGTTCAAAAGAAACTTGAAAAGGCTAAAAAATCCAAGAACGGTATTGAACAAACC ATTAACAGTAAGGATTACGAAACAAAGGCTAATACACAGGCCAAGGAAGCCAATAAAAGC AAGCTGGATAACACTGTTGCCGAAATCGAAGGTTTGGAAGCTACTATTGAAAACTTGAAG CGTTTGAAATTGTAG |
MNKWLNTLSKTFTFRLLNCHYRRSLPLCQNFSLKKSLTHNQVRFFKMSDLDNLPPVDPKT GEVIINPLKEDGSPKTPKEIEKEKKKAEKLLKFAAKQAKKNAAATTGASQKKPKKKKEVE PIPEFIDKTVPGEKKILVSLDDPALKAYNPANVESSWYDWWIKTGVFEPEFTADGKVKPE GVFCIPAPPPNVTGALHIGHALTIAIQDSLIRYNRMKGKTVLFLPGFDHAGIATQSVVEK QIWAKDRKTRHDYGREAFVGKVWEWKEEYHSRIKNQIQKLGASYDWSREAFTLSPELTKS VEEAFVRLHDEGVIYRASRLVNWSVKLNTAISNLEVENKDVKSRTLLSVPGYDEKVEFGV LTSFAYPVIGSDEKLIIATTRPETIFGDTAVAVHPDDDRYKHLHGKFIQHPFLPRKIPII TDKEAVDMEFGTGAVKITPAHDQNDYNTGKRHNLEFINILTDDGLLNEECGPEWQGMKRF DARKKVIEQLKEKNLYVGQEDNEMTIPTCSRSGDIIEPLLKPQWWVSQSEMAKDAIKVVR DGQITITPKSSEAEYFHWLGNIQDWCISRQLWWGHRCPVYFINIEGEEHDRIDGDYWVAG RSMEEAEKKAAAKYPNSKFTLEQDEDVLDTWFSSGLWPFSTLGWPEKTKDMETFYPFSML ETGWDILFFWVTRMILLGLKLTGSVPFKEVFCHSLVRDAQGRKMSKSLGNVIDPLDVITG IKLDDLHAKLLQGNLDPREVEKAKIGQKESYPNGIPQCGTDAMRFALCAYTTGGRDINLD ILRVEGYRKFCNKIYQATKFALMRLGDDYQPPATEGLSGNESLVEKWILHKLTETSKIVN EALDKRDFLTSTSSIYEFWYLICDVYIENSKYLIQEGSAIEKKSAKDTLYILLDNALKLI HPFMPFISEEMWQRLPKRSTEKAASIVKASYPVYVSEYDDVKSANAYDLVLNITKEARSL LSEYNILKNGKVFVESNHEEYFKTAEDQKDSIVSLIKAIDEVTVVRDASEIPEGCVLQSV NPEVNVHLLVKGHVDIDAEIAKVQKKLEKAKKSKNGIEQTINSKDYETKANTQAKEANKS KLDNTVAEIEGLEATIENLKRLKL |
The predicted MW for this protein is 58.21kD.
Nucelotide Sequence Alignment
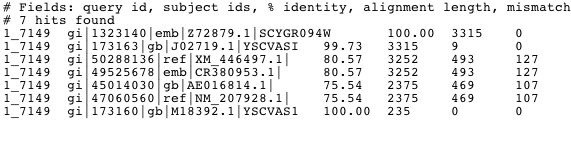
Figure 4. The image above shows the results of a megaBLAST on the sequence of Vas1. This shows signficant alignment to homologs of the Vas1 gene in other species including the fungi Eremothecium gossypii (NM_207928).
This Blastn search does not reveal much novel information about the Vas1 sequence. However, Xavier Jordana and collegues have shown that the sequence of Vas1 is 23% homologous to isoleucyl-tRNA synthetase in E.Coli. This similarity is the highest ever reported between genes of this family from different species and might be evidence of a close evolutionary relationship between the genes and/or organisms (1986).
Non-annotated Gene: YGR093W
Chromosomal Location
This candidate gene is located on Chromosome VII between bp 670392 and 671915, just upstream of Vas1 (Figure 1). Like Vas1, YGR093W seems use the top strand as the coding strand during transcription.
Biological Process
Currently Unknown
Molecular Function
Currently Unknown
Cellular Component
YGR093W seems to be expressed mainly in the nucleus of Yeast cells.
Nucleotide Sequence |
Predicted Amino Acid Sequence |
| ATGACAAATGCAAAGATTTTAGTAGCTCATATAAGTGAAAGCGATGCCGATGAGGCTATC AGAAAGATCAAGAAAGTGAATGAAAAATCAGGGCCCTTTGATCTAATAATTATATTCAGT AACTCGTATGATGAAAATTTTGAGCTGAATACTGATGGGTTACCTCAACTAATACTACTA TCGTGTGATAAGGCTAACAATTCGAAATCCAAAAAGATAAATGAAAATGTAACATTGCTG CATAATATGGGTACTTATAAATTAGCAAATGGAATCACTCTTTCATATTTTATTTATCCG GATGATACTCTTCAAGGGGAGAAAAAAAGCATACTGGACGAATTTGGCAAAAGTGAGGAT CAGGTAGACATTCTCCTTACAAAAGAATGGGGCCTTTCGATCTCTGAGAGATGTGGAAGG TTGTCTGGAAGTGAAGTTGTTGATGAATTGGCGAAAAAGTTACAAGCAAGGTACCATTTT GCCTTTTCAGATGAAATAAACTTTTACGAATTAGAGCCTTTCCAGTGGGAAAGAGAGCGC TTATCGAGGTTCCTCAATATTCCAAAATATGGATCTGGAAAGAAATGGGCCTATGCATTC AATATGCCAATAGGGGACAACGAACTAAAGGATGAACCTGAACCGCCCAACTTGATAGCT AACCCGTATAATAGCGTGGTTACAAACAGCAATAAAAGGCCACTAGAAACAGAAACAGAG AATTCGTTCGATGGAGACAAACAGGTACTTGCTAATAGAGAAAAGAATGAAAATAAAAAA ATTCGAACGATTTTGCCGTCAAGTTGTCATTTCTGCTTTTCAAATCCAAACCTCGAGGAT CATATGATAATATCAATCGGCAAACTAGTGTATTTAACCACAGCGAAGGGACCTTTAAGT GTTCCTAAGGGTGATATGGATATCTCAGGCCATTGCCTCATTATTCCCATTGAACATATT CCGAAATTAGATCCAAGCAAGAACGCAGAGTTGACACAGAGTATTTTGGCTTATGAAGCT AGTCTTGTGAAGATGAACTACATAAAATTTGATATGTGCACGATTGTCTTCGAAATACAG TCTGAACGTTCTATTCATTTCCACAAACAAGTTATTCCCGTTCCAAAATACCTCGTTCTA AAGTTCTGCAGTGCCTTAGATAGACAGGTTCATTTCAATAACGAAAAATTCACAAGAAAT GCTAAGCTAGAGTTCCAATGTTACGATTCACACTCTTCCAAACAATATGTGGATGTAATT AACAACCAATCCAATAATTATTTACAATTTACCGTCTACGAGACTCCTGAAGCGGACCCA AAGATATATTTGGCCACATTTAATGCCAGTGAGACAATAGATCTGCAGTTTGGACGACGT GTACTAGCCTTTTTACTTAACTTGCCACGCAGGGTGAAATGGAATTCTTCAACCTGTTTA CAAACTAAGCAACAAGAGACTATAGAGGCTGAAAAGTTTCAAAAGGCCTACAGGACCTAT GACATTTCTCTCACAGAAAACTAA |
MTNAKILVAHISESDADEAIRKIKKVNEKSGPFDLIIIFSNSYDENFELNTDGLPQLILL SCDKANNSKSKKINENVTLLHNMGTYKLANGITLSYFIYPDDTLQGEKKSILDEFGKSED QVDILLTKEWGLSISERCGRLSGSEVVDELAKKLQARYHFAFSDEINFYELEPFQWERER LSRFLNIPKYGSGKKWAYAFNMPIGDNELKDEPEPPNLIANPYNSVVTNSNKRPLETETE NSFDGDKQVLANREKNENKKIRTILPSSCHFCFSNPNLEDHMIISIGKLVYLTTAKGPLS VPKGDMDISGHCLIIPIEHIPKLDPSKNAELTQSILAYEASLVKMNYIKFDMCTIVFEIQ SERSIHFHKQVIPVPKYLVLKFCSALDRQVHFNNEKFTRNAKLEFQCYDSHSSKQYVDVI NNQSNNYLQFTVYETPEADPKIYLATFNASETIDLQFGRRVLAFLLNLPRRVKWNSSTCL QTKQQETIEAEKFQKAYRTYDISLTEN |
Predicted Protein Structure and Function
Using the website-based protein domain predictors PREDATOR and Conserved Domain, some hypotheses about the possible functions of this gene product can be made.
PREDATOR : Alpha helix (Hh) : 143 is 28.21% 310 helix (Gg) : 0 is 0.00% Pi helix (Ii) : 0 is 0.00% Beta bridge (Bb) : 0 is 0.00% Extended strand (Ee) : 77 is 15.19% Beta turn (Tt) : 0 is 0.00% Bend region (Ss) : 0 is 0.00% Random coil (Cc) : 287 is 56.61% Ambigous states (?) : 0 is 0.00% Other states : 0 is 0.00%
Figure 5. The graph above shows secondary and tertiary structure of the YGR093W protein along its predicted amino acid chain. The table over the graph provides a key for the color coding and the percent coverage of these regions over the entire protein chain.

Figure 6. The Conserved Domain website provides information on protein domains that align well to the provided amino acid sequence. YGR093W aligns very well to the proteins CwfJ_C_1 (E= 2e^-35) and CwfJ_C_2 (E=7e^-24).
Structural Conclusions
PREDATOR predicts that 28% of this protein will fold into an alpha-helices and 15% will remain as extended strands. However, the program predicts that over 56% of this protein will be "random coil". In other words, the program cannot reliably predict what the majority of this protein will look like in its fully folded (actual) form. This casts some doubt on the regions predicted to behave in a certain way. If the program cannot predict how over half of the protein will be shaped, how accurate are predictions about the rest of the protein. Unfortunately, we learn little about the structure of the YGR093W predicted protein from PREDATOR analysis. However, the Conserved Domain analysis of this amino acid sequence produces more useful results. Neighboring portions of the YGR093W amino acid sequence are very similar to the N-terminus of the proteins CwfJ_C_1 and CwfJ_C_2. The relatively low E values of these alignments show that they are reliable. These proteins are involved in an mRNA splicing complex in Schizosaccharomyces pombe, another species of yeast (Marchler-Bauer, 2005). Since the predicted N-terminus contains adjacent regions similar to the N-terminus of each of these proteins, it is likely that the genes have a related biological process or similar molecular functions. At the very least, these genes are related evolutionarily.
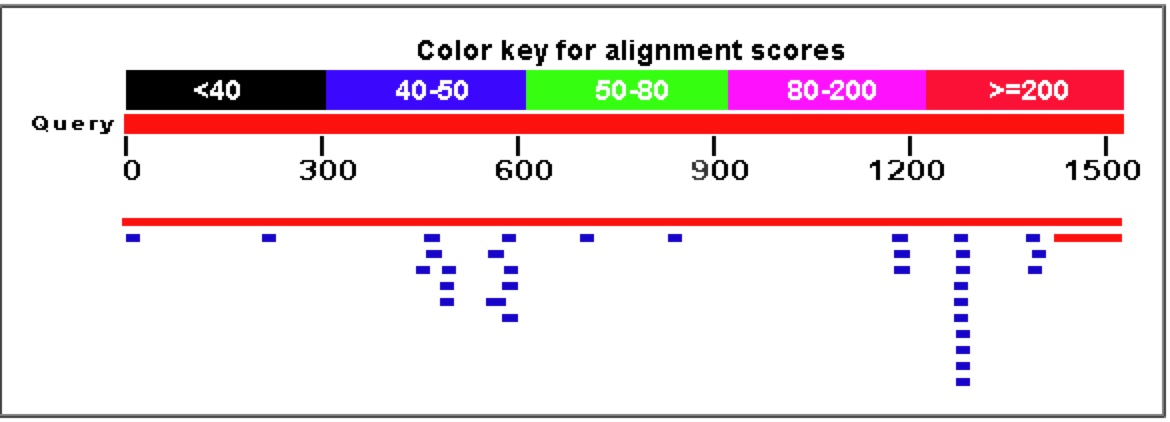
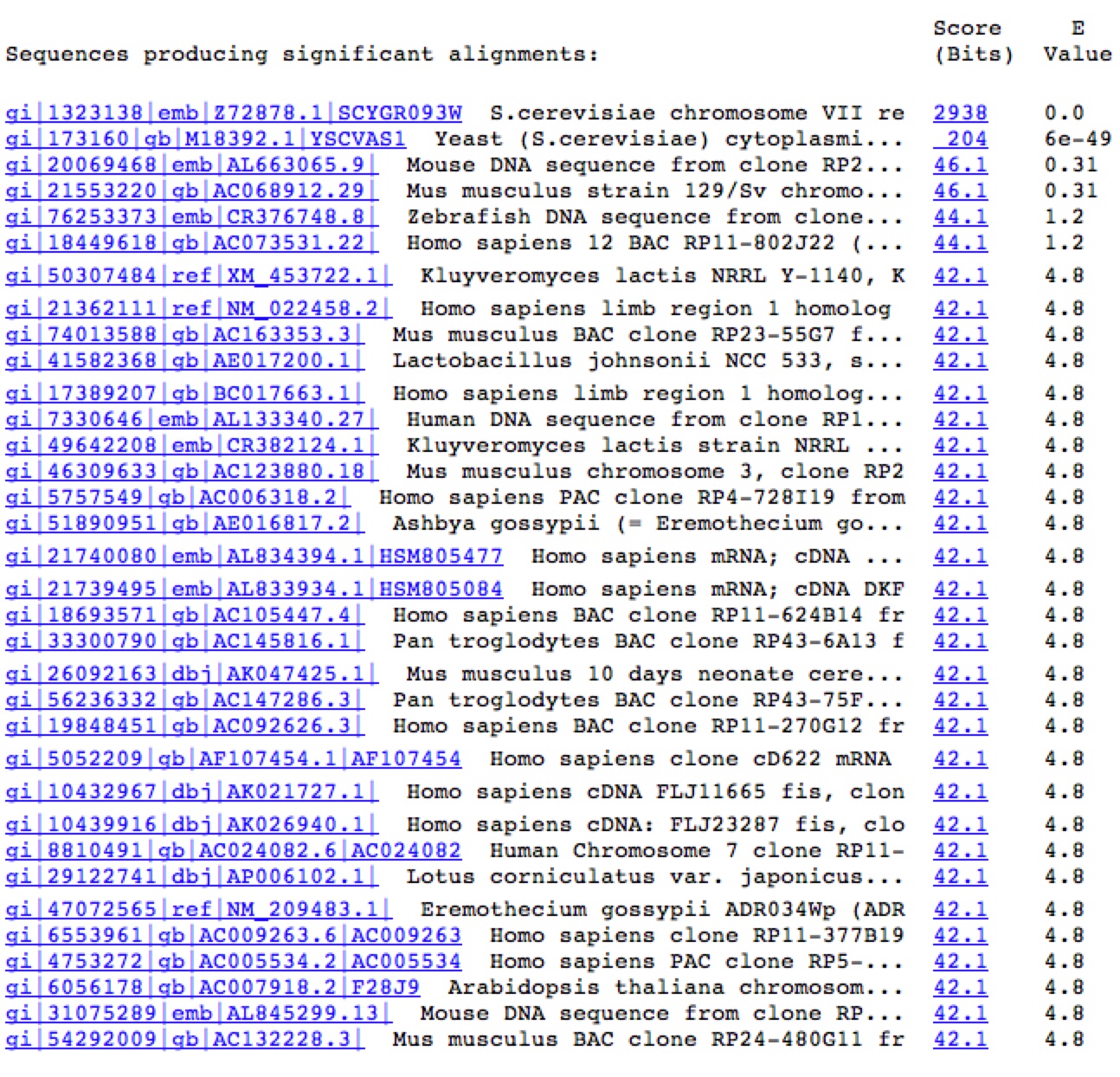
Figure 7. A Blastn search on the nucleotide sequence of YGR093W reveals very few similar nucleotide sequences. The only significant E values (top two rows) come from the sequence matching up to itself and the 5' end of YGR093W matching up to Vas1 where their genomic sequences overlap.
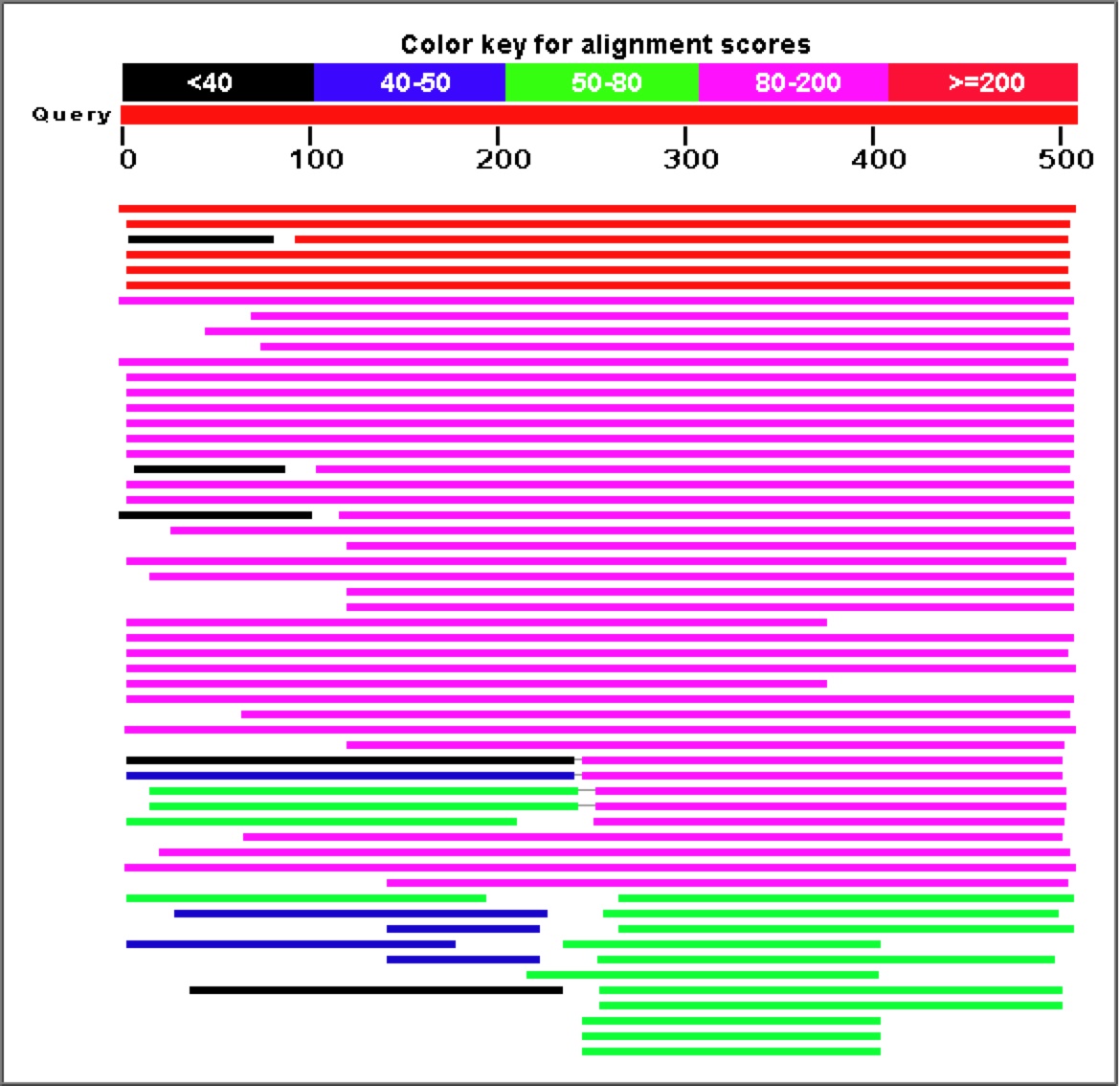

Figure 8. As is obvious from the graph and corresponding table above, a Blastp search for similar amino acid sequences to YGR093W reveals a large number of highly similar proteins.
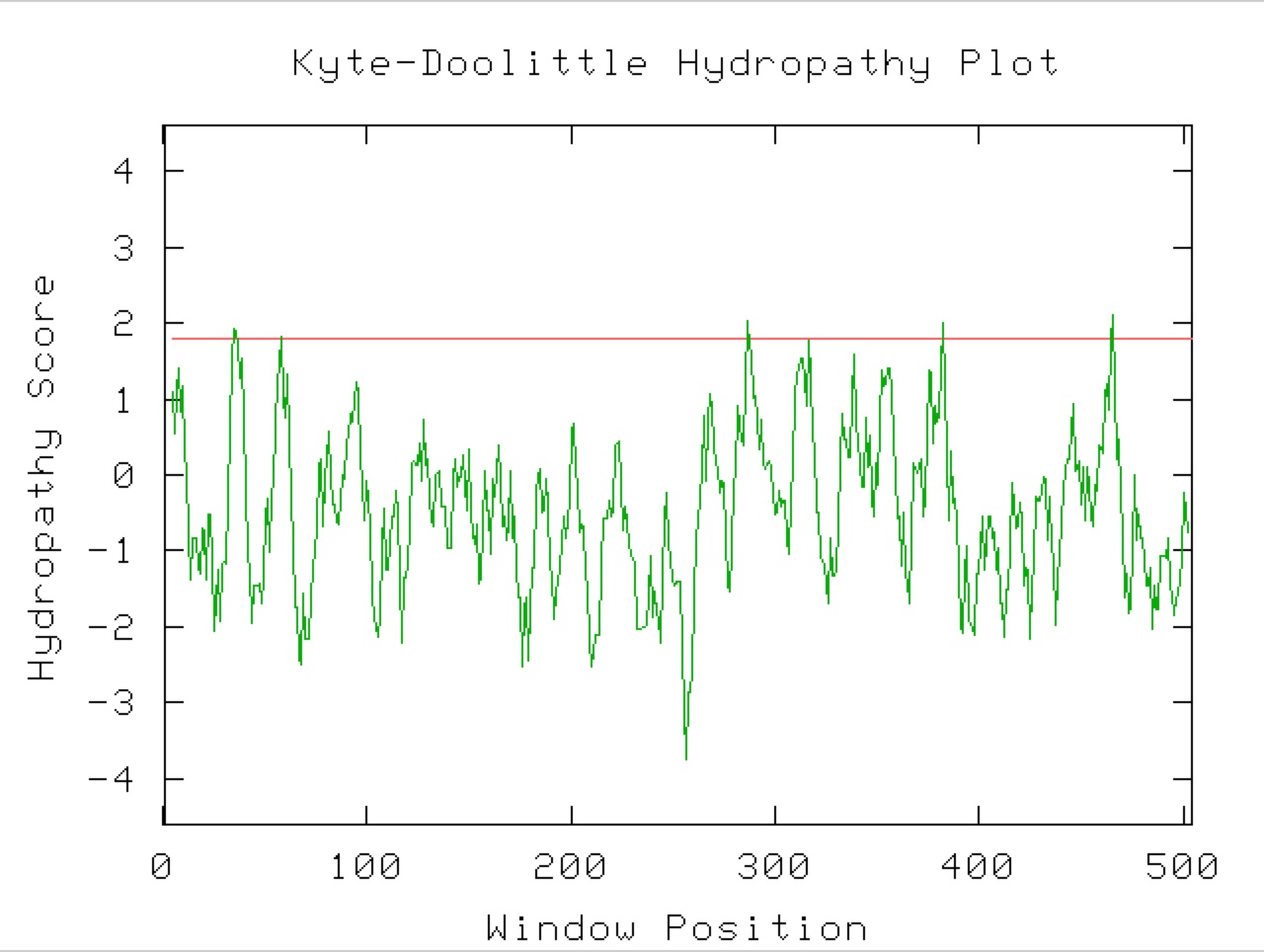
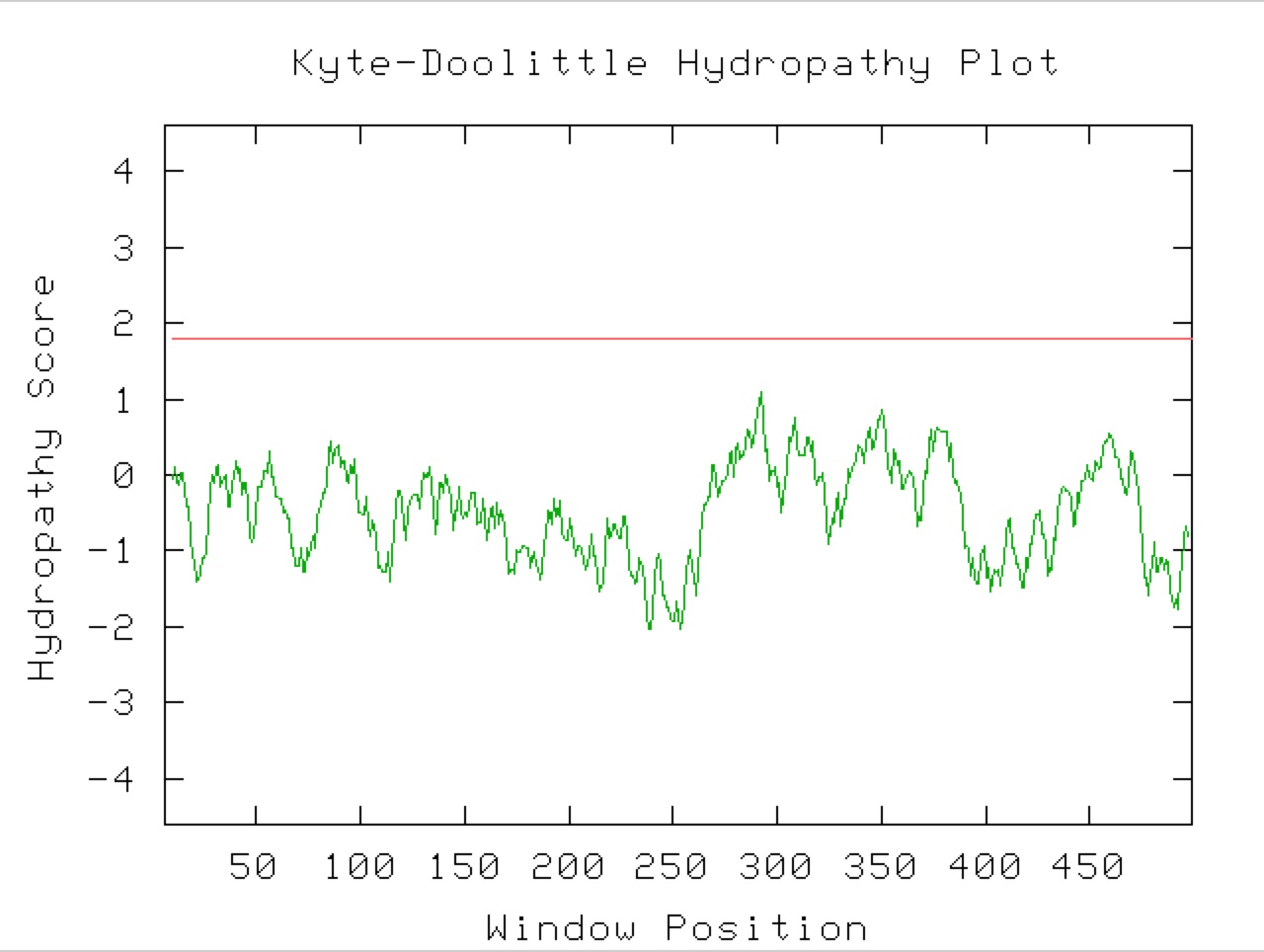
Figure 9. A Kyle-Doolittle Plot with a window size of 19 (right) shows no predicted transmembrane regions on the YGR093W predicted protein, evidenced by no regions that come close to crossing the red line. With a window size of 9 (left), the plot shows predicted surface portions of a globular protein. It is unclear exactly which sections of this protein would be at the surface since not many regions greatly exceed the specified limit (red line).
Functional Conclusions
Comparing the Blastn and Blastp results from the nucleotide and predicted amino acid sequence of YGR093W reveals some surprises. One might expect to see large similarities in the results of these two searches. However, YGR093 shows no significant similarity to any other genes while exhibiting much similarity to the primary sequences of many proteins. This striking result suggests that perhaps the mRNA transcripts that code for these similar proteins are spliced heavily before they are translated. Apart from several hypothetical proteins, YGR093W is similar to Cwf family proteins in mouse and the fungus Aspergillus fumigatus. These proteins seems to be highly conserved among divergent species. These proteins seem to take part in mRNA splicing somehow, but their molecular function is currently unknown. However, it has been shown that these proteins do form part of the spliceosome (Marchler-Baue et al,. 2005) . Since YGR093W protein shows conserved domains with proteins involved in mRNA splicing and exhibits much amino acid sequence similarity to proteins involved in spliceosomes, it is very likely that YGR093 codes for a protein that forms part of or interacts with the spliceosome in yeast.
References
Baldwin A N, and Berg P,. 1966. J Biol. Chem. 241: 839-842.
Chatton B, Walter P, Ebel J, Lacroute F, and Fasiolo F,. 1987. The Yeast Vas1 Gene Encodes Both Mitochondrial and Cytoplasmic Valyl-tRNA Synthetases. J Biol Chem. 261 (1): 52-57.
Jordana X, Chatton B,. Paz-Weisshaar M, Buhler J, Cramer F, Ebel J, and Fasiolo F,. 1986. Structure of the Yeast Valyl-tRNA Synthetase Gene (VAS1) and the Homology of Its Translated Amino Acid Sequence with Escherichia coli Isoleucyl-tRNA Synthetase. J Biol Chem 262 (15): 7189-7194.
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. 2005. "CDD: a Conserved Domain Database for protein classification.", Nucleic Acids Res. 33: D192-6. <http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi>. Accessed 2005 October 6.
Wikipedia. 2005 August 15. Aminoacyl tRNA Synthetase. <http://en.wikipedia.org/wiki/Aminoacyl_tRNA_synthetase>. Accessed 2005 October 3.
Links
Davidson College Biology Department
© 2005 Department of Biology, Davidson College, Davidson, NC 28036
Please direct comments, criticisms and questions to andrysdale "at" davidson.edu