This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Genes
Annotated gene: GDA1
GDA1 is expressed in Golgi vesicles of Saccharomyces cerevisiae. The gene produces guanosine diphosphate (GDPase) within the Golgi. GDPase converts GDP to GMP, which then allows GDP-mannose to enter the lumen of the Golgi vesicles. Within the vesicles, GDP-mannose acts as a substrate for mannosylation of glycoproteins and glycosphingolipids. Without the presence of GDPase, mannosylation of proteins and lipids within the Golgi vesicles would be severely impaired (Berninsone, Miret & Hirschberg, 1994).
Location
GDA1 resides on the fifth chromosome of S. cerevisiae at coordinates 73771 to 75327 (Figure 1) (SGD, 2005; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YEL042W). To view the complete gene sequence, click here.
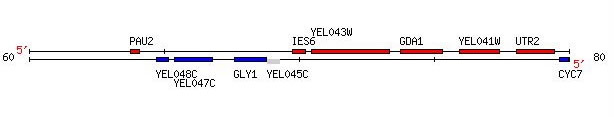
Figure 1. GDA1 is flanked by two non-annotated genes, YEL043W, which is discussed later in this web site, and YEL041W. Image provided by http://db.yeastgenome.org. Permission pending.
Function of GDA1
Biological Process: GDA1 is involved the biological process protein amino acid glycosylation, during which sugars are added to proteins. So far, eighteen other yeast genes have been identified as contributors to protein amino acid glycosylation (SGD, 2005; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=GDA1).
Biological Function: As mentioned above, GDA produces GDPase. GDPase combines with water within the Golgi to form GMP, which then permits GMP-mannose to enter the lumen and catalyze mannosylation. A phosphate group also results from this interaction. GDA is also involved in uridine diphosphate activity, although S. cerevisiae does not use uridine sugars within its Golgi lumen. In this second case, UDP combines with water to form UMP and a phosphate (Lopez-Avalos MD et al., 2001).
Cellular Component: Gda1p, a protein expression of GDA1, is located within the Golgi apparatus. Proteins are modified and processed within the Golgi. Processing includes sorting proteins for transport throughout the cell. Modifications include addition of sugar units onto glycoproteins, explaining the presence of Gda1p proteins within the Golgi (SGD, 2005; http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=5794).
Mutant Alleles and Phenotypes
GDA1 exhibits seven recorded mutant phenotypes. Only two of the mutations produce viable cells, one of which is a null mutation, the other a systematic deletion. Two other null mutations cause a defect in Gdap1's ability to mannosylate proteins and sphingolipids. One of these mutant alleles, designated gda1, produces mutant vesicles that experience a decreased rate of GDP-mannose entry into the cell. This causes impaired mannoprotein synthesis, and therefore altered mannosylation of proteins and lipids (Berninsone et al., 1994). Another systematic deletion causes a five generational sensitivity to a weakly basic environment. The two remaining mutant phenotypes result from homozygous systematic deletions, one of which causes reduced fitness in a rich medium and the other which causes an increased accumulation of glycogen within S. cerevisiae (SGD, 2005; http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?dbid=S000000768). For more information on these mutant phenotypes, click here.
Gdap1
Gdap1 is 518 amino acids long, has a MW of 56.8 kilodaltons (SGD, 2005; http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000000768). To view its sequence, click here.
Gdap1 is recorded as physically interacting with five other proteins (SGD, 2005; http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?dbid=S000000768&type=allinteractions#physical). These interactions are classified as two-hybrid interactions (Figure 2). To view a list of proteins included in these interactions, click here.
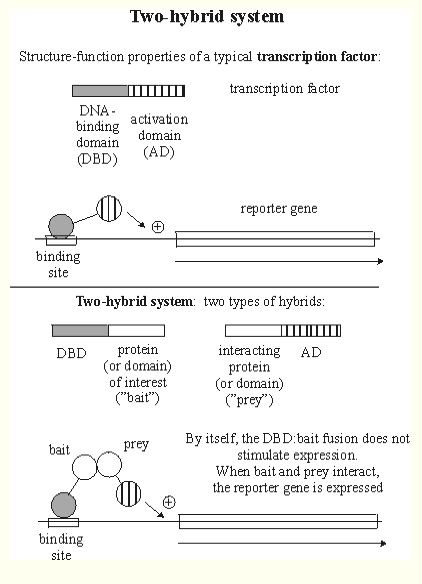
Figure 2. This is a graphical explanation of two-hybrid protein interactions. The DNA-binding domain (DBD) hybrid binds to the "bait" (protein of interest), but cannot activate transcription. However, after binding with the activation domain (AD), both genes are expressed and the protein of interest can be isolated using the reporter gene as a marker. Image from http://www.biochem.arizona.edu/classes/bioc568/two-hybrid_system.htm. Permission pending.
Gdap1 is also involved in an synthetic lethality interaction with protein arl3. Synthetic lethality refers to a condition where a single mutation in one gene or the other produces viable cells, but both mutations result in inviable cells (SGD, 2005; http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/rev-sup/synthetic.html). For more information of synthetic lethality, click here.
A Conserved Domain search returned hits for GppA and Ppx-GppA, proteins involved in nucleotide and inorganic ion transport and metabolism (Figure 3) (NCBI, 205; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi).
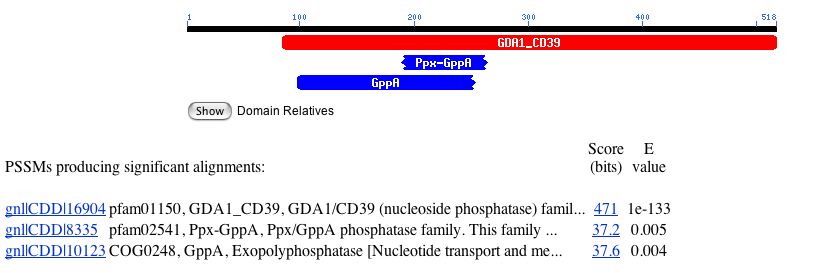
Figure 3. Results from a Conserved Domain query of Gdap1. Image from http://www.ncbi.nlm.nih.gov/. Permission pending.
A homology search of GDA1 returned many hits (Figure 4). This list suggests that a version of the GDA1 gene exists in many other organisms. Since GDA1 has a fundamental role of modifying proteins within the Golgi, this result is not surprising.
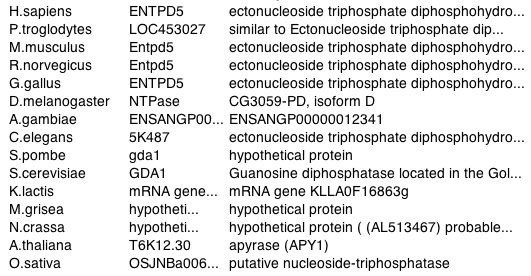
Figure 4. List of organisms with GDA1 homologous gene. Image courtesy of http://www.ncbi.nlm.nih.gov/entrez/. Permission pending.
According to Figure 5, Gdap1 contains a transmembrane domain (SDB, 2005; http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000000768).
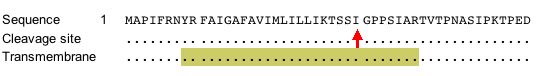
Figure 5. This image shows the portion of amino acid sequence that defines the protein as transmembrane. Image from http://db.yeastgenome.org. Permission pending.
A Kyte-Doolittle Hydropathy plot also suggests Gdap1 contains a integral membrane region (Figure 6).
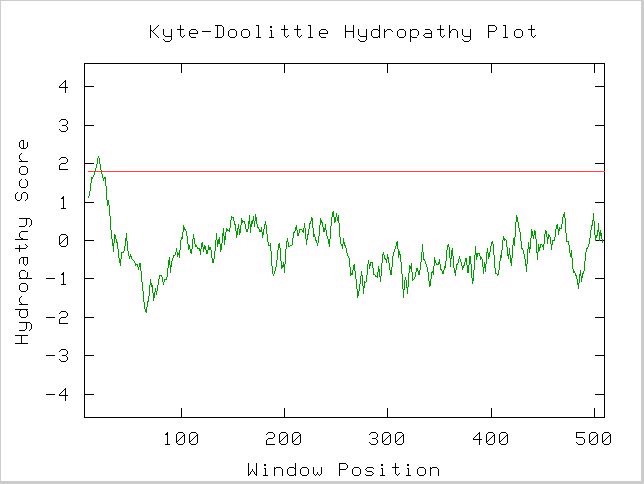
Figure 6. A spike that crosses the red line signals the possibility of a transmembrane protein. Image obtained using protein sequence available at link above. http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/slrkd.pl was used to create image.
Structure of Gdap1
The structure of a protein can greatly influence its function and protein-protein interactions. The above Kyte-Doolittle plot suggests a transmembrane domain within the protein.
Figure 7 predicts the secondary structure of Gdap1 using possible hydrogen bonds gathered from its amino acid sequence.
Figure 7. Proposed secondary structure of Gdap1. Blue indicates alpha-helices, red indicates regions of extended strands, and purple sections suggest portions of random coil. Image courtesy of http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl#preda. Permission pending.
Non-annotated Gene: YEL043W
Location
YEL043W is an non-annotated gene adjacent to GDA1, as seen in Figure 8 (SDB, 2005; http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000000769) .
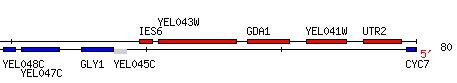
Figure 8. Notice YEL043W directly to the left of GDA1 on the Watson strand of chromosome V of Saccharomyces cerevisiae. YEL043W coordinates are 60478 - 83348. Image courtesy of http://db.yeastgenome.org/. Permission pending.
Function of YEL043W
The function of YEL043W is unknown at this point. However, examining its structure may provide us with hints about its function in S. cerevisiae.
The gene YEL043W has an ORF 2,870 bp long. To view the complete sequence, click here. The YEL043W protein is 956 amino acids long and has a molecular weight of 106.132 kilodaltons. To view the amino acid sequence, click here (SDB, 2005; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YEL043W).
Although the function of YEL043W is yet to be determined, it does undergo two synthetically lethal interactions, both resulting in inviable cells. Four protein-protein interactions of YEL043W were documented during a comprehensive study examining two-hybrid protein interactions within Saccharomyces cerevisiae (SDB, 2005; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YEL043W). To see a list of these interactions, click here.
Performing a BLASTp of the amino acid sequence of YEL043W returns the following list of proteins with relatively similar sequences (similarity quantified by the e-value) (Figure 9).

Figure 9. A portion of the hits from a BLASTp query of the amino acid sequence of YEL043W. Image from http://www.ncbi.nlm.nih.gov/blast/Blast.cgi. Permission pending.
Searching the PDB Homolog database returns a hit for a protein within Gallus gallus (Figure 10).
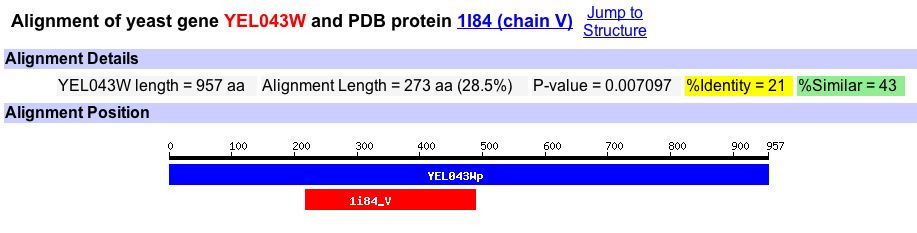
Figure 10. An image from PDB Homology that reports the percent identity and percent similarity between S. cerevisiae gene YEL043W and G. gallus protein 1I84. This is the only homology hit for YEL043W. Image from http://db.yeastgenome.org/. Permission pending.
The G. gallus protein 1I84 is conserved within a family of proteins involved in forming a myosin tail within eukaryotic cells. The proteins have tight alpha-helix secondary structure to enable contraction of the cell. Although the alignment score of 1I84 within this family is only 19.1%, this may provide some clues to the function of YEL043W (NCBI, 2005; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi)
Structure of YEL043W
Similar to GDA1, YEL043W has a transmembrane domain, although it only includes 17 amino acids within the protein (Figure 11) (SDB, 2005; http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1i84).
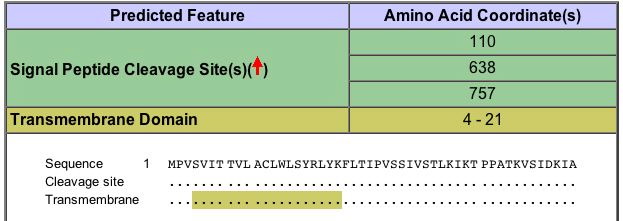
Figure 11. Image highlighting the transmembrane domain of YEL043W ORF. Image courtesy of http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1i84. Permission pending.
A Kyte-Doolittle plot shows a peak in the same area (amino acids 4 through 21). The peak is somewhat weak compared to the ones observed within GDA1, but it may still possess a transmembrane domain (Figure 12).
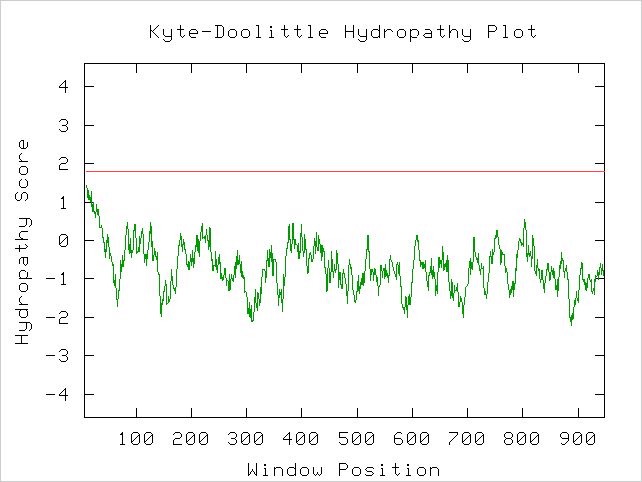
Figure 12. Kyte-Doolittle hydropathy plot of the ORF YEL043W. Peaks that cross the red line signal transmembrane protein behavior. It is unclear whether this protein is transmembrane or not. Image obtained using http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/slrkd.pl .
PREDATOR makes the following predictions about the secondary structure of ORF YEL043W (Figure 13).
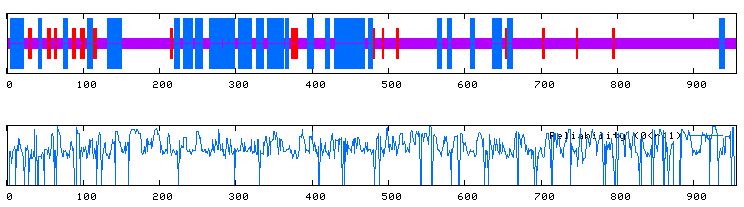
Figure 13. The top image predicts the secondary structure of ORF YEL043W. Blue indicates alpha-helices, red indicates regions of extended strands, and purple sections suggest portions of random coil. The bottom image displays the reliability of the predictions of the top image. Image courtesy of http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl. Permission pending.
Predictions
Based on prevalence of alpha-helices from the PREDATOR prediction, and the homology of YEL043W with the Kluyveromyces lactis gene 0F16819g, I predict the ORF YEL043W is involved in maintaining the cytoskeletal structure of cells within S. cerevisiae. The alpha-helices suggest this protein is strong, and the homology with a myosin family offers a possible lead for YEL043W's involvement in cellular cytoskelton.
References
Berninsone, P, Miret, J, Hirschberg, C. 1994. The Golgi guanosine diphosphatase is required for transport of GDP-Mannose into the lumen of Saccharomyces cerevisiae Golgi vesicles. <http://www.jbc.org/cgi/reprint/269/1/207>. Accessed 2005 Oct 4.
Kyte Doolittle Hydropathy Plot. <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm> Accessed 2005 Oct 4.
Leung, Y. (2004). Functional Genomics. <http://ihome.cuhk.edu.hk/%7Eb400559/array.html>. Accessed 2005 Oct 21.
Little, J. 2004. Two-hybrid system for predicting protein-protein interactions. <http://www.biochem.arizona.edu/classes/bioc568/two-hybrid_system.htm>. Accessed 2005 Oct 4.
Lopez-Avalos, M, Uccelletti, D, Abeijon, C, Hirschberg, C. 2001. The UDPase activity of the Kluyveromyces lactis Golgi GDPase has a role in uridine nucleotide sugar transport into Golgi vesicles. <http://glycob.oxfordjournals.org/cgi/content/full/11/5/413>. Accessed 2005 Oct 5.
NCBI (National Society for Biotechnology Information). 2005.<http://www.ncbi.nlm.nih.gov/>. Accessed 2005 Oct 4.
PREDATOR. 2005. <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html>. Accessed 2005 Oct 4.
SGD (Saccharomyces Genome Database). 2005. < http://www.yeastgenome.org/ >. Accessed 2005 Oct 4.
Questions or comments? E-mail Caitlin Kiley at cakiley@davidson.edu
Return to Davidson College Biology Home Page
Return to Genomics Home Page
Return to Caitlin Kiley's Home Page
Return to Davidson College Home Page