This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Genes: SAE2 and YGL176C
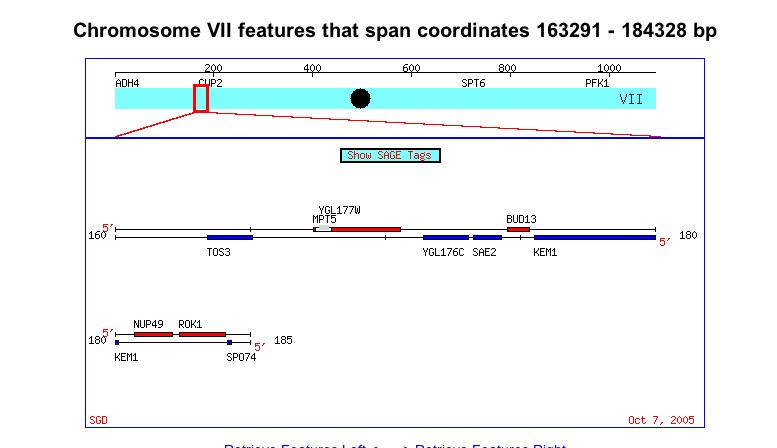
Chromosomal Location: Yeast genes SAE2 and YGL176C are located on chromosome VII between coordinates 160,000 and 180,000. These genes are positioned directly beside each other, on the Crick-strand (Figure 1).

Figure 1. Screen Shot of Chromosome VII. Notice YGL176 and SAE2 in blue (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000003143>.
Annotated Gene: SAE2
Referred to in the literature as COM1 and systematically named YGL175C, SAE2 encodes a DNA repair enzyme.
DNA Information
The genomic DNA of this gene spans coordinates 174328 and 173291 and sequence in GCG format is as follows:
1 ATGGTGACTG GTGAAGAAAA TGTGTATCTA AAGTCAAGCT TATCCATTCT
51 CAAGGAGCTC AGTCTCGATG AACTACTAAA TGTGCAGTAT GACGTGACTA
101 CCTTAATCGC GAAACGTGTT CAAGCACTGC AAAATCGGAA TAAATGTGTT
151 CTCGAAGAAC CTAACAGTAA ACTGGCTGAA ATTCTATGTC ACGAAAAAAA
201 TGCTCCTCAA CAATCCTCTC AGACGTCTGC GGGGCCAGGT GAGCAAGATT
251 CTGAAGATTT CATCCTTACT CAGTTTGATG AGGACATAAA GAAAGAATCA
301 GCAGAGGTTC ACTATCGTAA TGAAAATAAA CACACTGTGC AGTTACCGCT
351 GGTTACTATG CCACCTAATA GGCATAAACG GAAAATTTCG GAGTTTAGTT
401 CGCCTTTAAA TGGCCTCAAC AACTTAAGCG ATTTGGAAGA TTGCTCTGAT
451 ACTGTAATCC ACGAAAAAGA TAATGATAAA GAGAACAAAA CAAGAAAACT
501 ATTGGGAATA GAGCTCGAGA ACCCTGAATC TACATCGCCA AATTTATACA
551 AAAATGTTAA GGATAATTTC CTGTTTGACT TCAATACGAA CCCTTTAACA
601 AAGCGAGCCT GGATTCTCGA AGACTTTAGA CCAAATGAAG ATATCGCCCC
651 GGTTAAGAGA GGAAGAAGAA AATTGGAGCG ATTTTATGCC CAAGTTGGAA
701 AGCCAGAGGA TTCTAAACAC AGATCATTGT CAGTAGTTAT AGAATCGCAA
751 AATTCGGACT ACGAATTTGC GTTTGATAAC TTGAGGAATA GATCAAAATC
801 CCCCCCAGGT TTTGGAAGAC TGGATTTTCC CTCCACTCAG GAAGGGAACG
851 AGGACAAAAA GAAATCCCAG GAAATCATCA GAAGAAAGAC AAAATATAGG
901 TTTTTAATGG CAAGCAATAA CAAGATCCCA CCGTACGAGA GAGAATATGT
951 ATTCAAAAGA GAACAATTAA ATCAAATTGT TGACGATGGA TGTTTCTTCT
1001 GGAGTGATAA ATTATTGCAG ATATATGCTA GATGTTAA
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/getSeq?map=amap&seq=YGL175C&flankl=&flankr=&rev=>).
By performing a BLASTn search, I found that the nucleotide sequence for SAE2 only has high similarity to itself. The sequence shows some similarity to small fragments of DNA from zebra fish and mice genomes, but the corresponding E-values (probability that the similarity occurred by chance) are high.
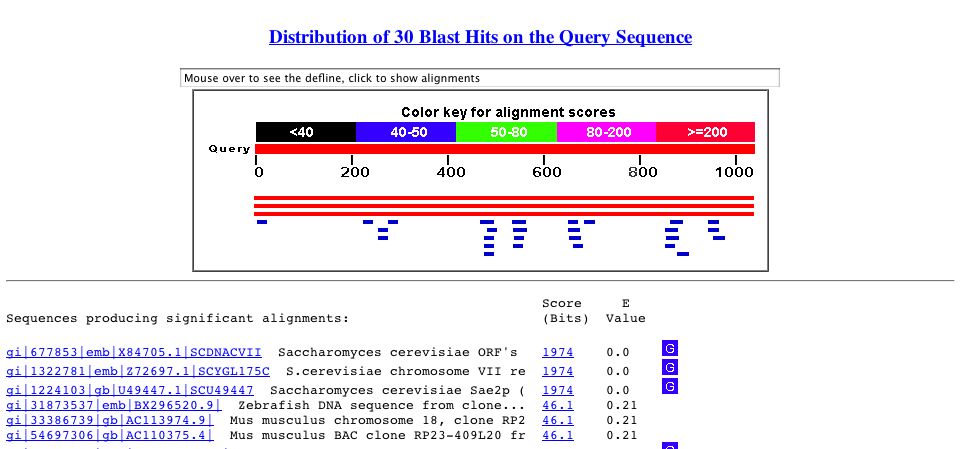
Figure 2. Blastn search results. Perform your own search by copying the sequence above and accessing Blastn.
The coding sequence, on the other hand , is as follows in GCG format:
1 ATGGTGACTG GTGAAGAAAA TGTGTATCTA AAGTCAAGCT TATCCATTCT
51 CAAGGAGCTC AGTCTCGATG AACTACTAAA TGTGCAGTAT GACGTGACTA
101 CCTTAATCGC GAAACGTGTT CAAGCACTGC AAAATCGGAA TAAATGTGTT
151 CTCGAAGAAC CTAACAGTAA ACTGGCTGAA ATTCTATGTC ACGAAAAAAA
201 TGCTCCTCAA CAATCCTCTC AGACGTCTGC GGGGCCAGGT GAGCAAGATT
251 CTGAAGATTT CATCCTTACT CAGTTTGATG AGGACATAAA GAAAGAATCA
301 GCAGAGGTTC ACTATCGTAA TGAAAATAAA CACACTGTGC AGTTACCGCT
351 GGTTACTATG CCACCTAATA GGCATAAACG GAAAATTTCG GAGTTTAGTT
401 CGCCTTTAAA TGGCCTCAAC AACTTAAGCG ATTTGGAAGA TTGCTCTGAT
451 ACTGTAATCC ACGAAAAAGA TAATGATAAA GAGAACAAAA CAAGAAAACT
501 ATTGGGAATA GAGCTCGAGA ACCCTGAATC TACATCGCCA AATTTATACA
551 AAAATGTTAA GGATAATTTC CTGTTTGACT TCAATACGAA CCCTTTAACA
601 AAGCGAGCCT GGATTCTCGA AGACTTTAGA CCAAATGAAG ATATCGCCCC
651 GGTTAAGAGA GGAAGAAGAA AATTGGAGCG ATTTTATGCC CAAGTTGGAA
701 AGCCAGAGGA TTCTAAACAC AGATCATTGT CAGTAGTTAT AGAATCGCAA
751 AATTCGGACT ACGAATTTGC GTTTGATAAC TTGAGGAATA GATCAAAATC
801 CCCCCCAGGT TTTGGAAGAC TGGATTTTCC CTCCACTCAG GAAGGGAACG
851 AGGACAAAAA GAAATCCCAG GAAATCATCA GAAGAAAGAC AAAATATAGG
901 TTTTTAATGG CAAGCAATAA CAAGATCCCA CCGTACGAGA GAGAATATGT
951 ATTCAAAAGA GAACAATTAA ATCAAATTGT TGACGATGGA TGTTTCTTCT
1001 GGAGTGATAA ATTATTGCAG ATATATGCTA GATGTTAA
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/getSeq?map=nmap&seq=YGL175C&flankl=&flankr=&rev=>).
The length of the genomic DNA is the same as the length of the coding sequence. Thus, it does not appear that SAE2 contains any introns.
Protein Information
The protein encoded by SAE2 has the following primary structure of amino acids:
Sequence 1 M V T G E E N V Y L K S S L S I L K E L S L D E L L N V Q Y D V T T L I A K R V Q A L Q N R N K C V
Sequence 51 L E E P N S K L A E I L C H E K N A P Q Q S S Q T S A G P G E Q D S E D F I L T Q F D E D I K K E S
Sequence 101 A E V H Y R N E N K H T V Q L P L V T M P P N R H K R K I S E F S S P L N G L N N L S D L E D C S D
Sequence 151 T V I H E K D N D K E N K T R K L L G I E L E N P E S T S P N L Y K N V K D N F L F D F N T N P L T
Sequence 201 K R A W I L E D F R P N E D I A P V K R G R R K L E R F Y A Q V G K P E D S K H R S L S V V I E S Q
Sequence 251 N S D Y E F A F D N L R N R S K S P P G F G R L D F P S T Q E G N E D K K K S Q E I I R R K T K Y R
Sequence 301 F L M A S N N K I P P Y E R E Y V F K R E Q L N Q I V D D G C F F W S D K L L Q I Y A R C *
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000003143>).
By performing a Blastp search, I found that the protein sequence is similar to itself and unnamed protein in other species of yeast, such as Candida glabrata and Kluyveromyces lactis. The protein produce to SAE2 seemed to be conserved across many species of yeast.
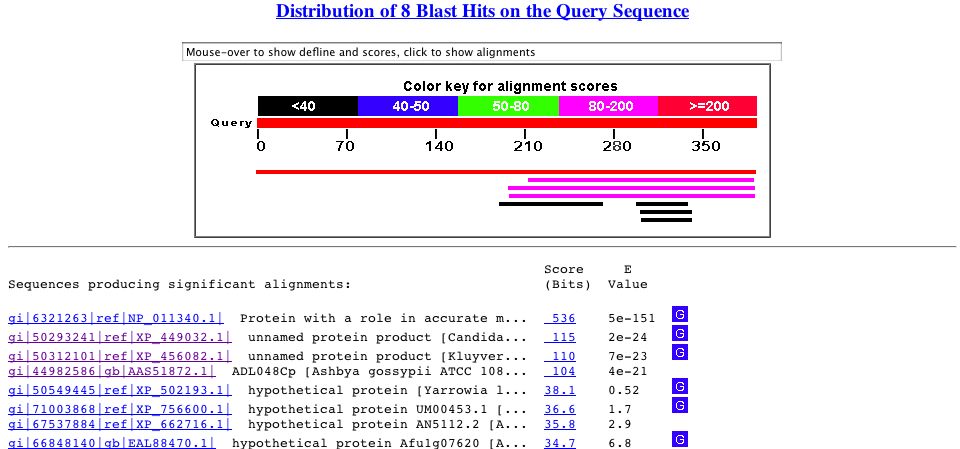
Figure 3. Blastp search results. Perform a search yourself by copying the protein sequence above and accessing Blastp.
Homologs:
Indeed, by searching the NCBI Homologene database, I found that SAE2 is conserved within yeast (Fig 4). The two homologs of SAE2 are more similar when comparing positives than simply identities.

Figure 4. Sceenshot of HomoloGene search results. Perform your own search for SAE2 at the NCBI Homologene website.
Molecular Function:
According to the SGD, the molecular function for SAE2 is unknown (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SAE2>).
Biological Process:
The protein encoded by SAE2 has been implicated in meiotic DNA repair (Roeder GS, 1997). The SAE2 protein shows 5'-3' exonuclease activity, meaning it has the ability to chew off DNA in the 5'-3' direction whenever DNA breaks (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=706>). This exonuclease activity is critical first set in the process of DNA repair (Fig 5).
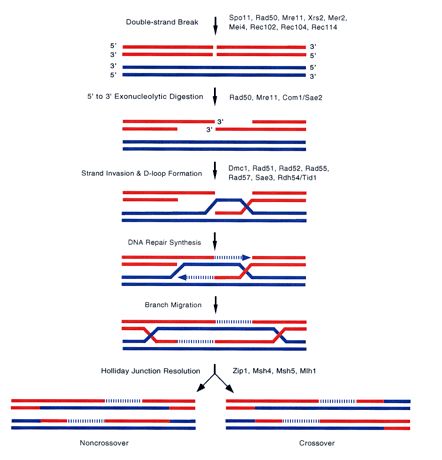
Figure 5. The process of yeast DNA break repair. Genes involved in the steps are listed next the the arrows. Notice SAE2/COM1 appears at the second arrow from the top. (Image from: Roeder GS., 1997, permission pending)
The SGD also describes SAE2 as having a role in mitotic DNA double-strand break repair (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SAE2>). Though crossing-over generally occurs in meiosis, homologous recombination also rarely occurs in mitosis. Rattray AJ, et al. (2001) assert that in mitosis, the protein product of SAE2 may help prevent break-induced replication by "ensuring both ends of a double-stranded break" participate in recombination event.
Cellular Component:
Scientists found that the protein transcribed by SAE2 in the cytoplasm and the nucleus with GFP (Huh WK, et al., 2003). Since chromosomes are located in the nucleus of eukaryotes, It makes sense the SAE2 protein product would be found in the nucleus given its proposed function in DNA break repair.
Mutant Alleles and Phenotypes:
The null-mutant has shown to be viable for SAE2 (Giaever G, et al., 2002). In other words, SAE2 knockout yeast can survive and SAE2 does not represent an essential yeast gene. Other genes must be present that function similarly to SAE2, so that yeast can survive without the SAE2 gene. According the the SGD (no alternative citation available), although viable, the null mutant is "weakly sensitive to methyl methane sulfonate, shows blocked turnover of meiosis-specific double-strand breaks, similar to rad50S mutant" (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YGL175C>). Methyl methane sulfonate is an alkylating agent that induces mutations in DNA by adding alkyl chains to bases.
Non-Annotated Gene: YGL176C
Located directly next to SAE2 on the Crick strand, YGL176 is a suspected gene (i.e. its sequence contains an open reading frame) of unknown function. The SDG lists this gene's molecular function, biological process, and cellular component as unknown.
DNA Information
The genomic DNA of this gene spans coordinates 173085 and 171421 and sequence in GCG format is as follows:
1 ATGACACCTA ACTTACCAGG GTTTTATTAT GATAGAGAAC GTCGACGTTA
51 TTTTCGCATA TCTGAAAATC AAAGCATTTC TACCACGGGA ACGACGAACC
101 AGTACAGAAA AGATAATATA AAGCGACAAT GTGTTGAGGA AAATTACGAT
151 AAAAAATTTT CTATGATTAA AAAAAAACGT CAGCAAACAT TACAAAAGTA
201 CAAGCTTAGT CTTCTGAATC CGTTGGAAAG GGCTTTTCGC CCTTTATCAT
251 ATGAAAAGTA CATGATTGGT TTAAATATGC AGTATGCATC GCATTCTCTC
301 ACTGAAGGTC ATCACTCACA TAGTTCTGCT AACGTAAAAT CACTCAATTT
351 TCCACACAGG ATACAGATAG GAGTATTAGC AAATTGTATT TTATTGGTAA
401 CTCAAGAAGG ATGTTTTCAT AGTAAATTAG TATTTGCAAC AAACAAGGGC
451 TATGTCGCAG GATTTTCATC CCTGGATAAT TTTTCTGAAG AAAATTTCTT
501 TACGGGGTTT TCCATGGCTG AATTAAATCC TATGTTGAAA TATAAGTCCG
551 AGCCAACTGA TGTGTTCAAA ACAATGAAAC TTGAGAGAAC AGTTGCCATC
601 AAGGAAGGGC CGAGCCATTA TTTTTATCAC AACGTTAATA CCAGATCTAA
651 CGTGCATACT TTTGCCATCT TTTTACAAGA CTTTTCTTCC TTAAAACTGC
701 TCAAAATTCG TCAGGTCAAA CTCAAAGAAA ATTGCCAAGT CCATGATTCT
751 CTTGTTGTTG GAGACACGTT GATTATTACC GTAAATTATC GTTGTCATTT
801 TTATGATTTG ATTCCAGAAA CCTTTCCAAA TCCCTATATC TTTTCTCCTG
851 CCAAAAGCTC AAGAAAACAC AAAAGTAGGA GTGACATTAC TTCCCTATCA
901 TTTTGTCTAC AAGAAGATGC TTTATCACCT TTAAAAAAAA CGAACACCGG
951 TGTGTTTTAT TTAGGTTACA GAAATGGTGA TTCAATGGCA ATAGTCTTCA
1001 CTAATATTAC AAACATGACA TTACAGTACT CCAAGACCAA CGGTATGACA
1051 TCAGAAAGTC GAAATCAACC TATCAGAAAT TCCCTAAAAT CTGTTGTTTC
1101 CATAAAAGCA TTGAATAATA AAGGCCTAAT ACTTATATCT GGAATGGCAG
1151 ACAAAGAGAA TGTGCAACAA CTGGTTATAG CAGATACGTT CCTGGAGGAC
1201 ATTCTGACCG AAATCCCGGT TGTATCTTTT AAAACTAAGT TCTTAAACGT
1251 GACTAAGGAT ACAGAAATAC TCGAGATATC GGATGACGGG CGCTACTTCA
1301 TTTATGGTTC AACTAGCGCT AGGGACGGAA AGGGTGATTT TGAAGTATTT
1351 TGTACCACTT TATCTGGAAA TCTTGACTAC GAAAAATCCG AAGGTGGAAA
1401 TATCACCCTC TATCCAATAG GAGGTATGAA GAACTACTGT AGGTTAGAAA
1451 ACTTCCAGTT TGAATCTATC CACCTACATT CGGCTTTCAT ACCCCCAAGG
1501 TACGTAAATC CGTTTGATGC TGTGGAACCT CTAGGTGAAG AGAGCAGTAC
1551 TTCACCTTAC GATATACCTG AAGAAGCACT TTCTCAAAAA ATATGCATCT
1601 TAATTCGGAG AGAAGATGAT CCCTACAATG GGGCCAATAT CTTCATAACA
1651 TCTGCTTTGA CCTAG
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/getSeq?map=amap&seq=YGL176C&flankl=&flankr=&rev=>).
By performing a BLASTn search, I found that the nucleotide sequence for YGL176C is most like itself (Fig 6-red region). However, this sequence shows similarity to a portion of the SAE2 gene (Fig 6- pink region).
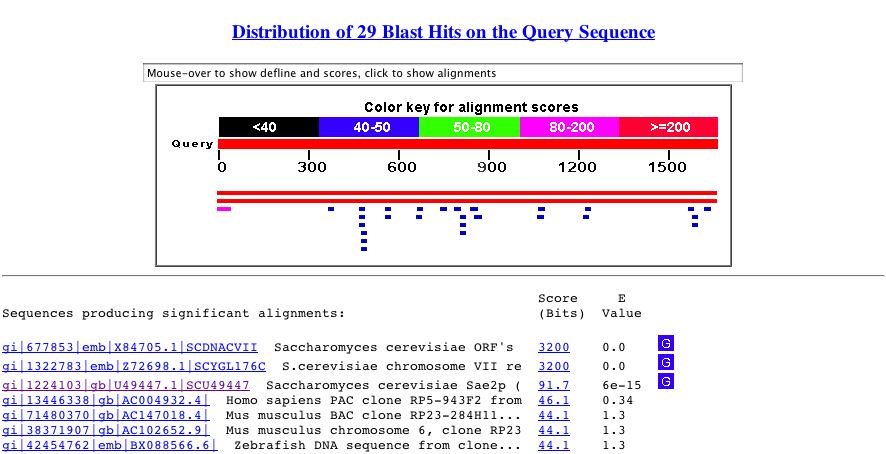
Figure 6. Blastn search results for YGL176C nucleotide sequence. Perform your own search by copying the sequence above and accessing Blastn.
Perhaps this similarity results because the open reading from of YGL176C overlaps with part of the SAE2 nucleotide sequence, as they are located directly next to each other.
The coding sequence, on the other hand , is as follows in GCG format:
1 ATGACACCTA ACTTACCAGG GTTTTATTAT GATAGAGAAC GTCGACGTTA
51 TTTTCGCATA TCTGAAAATC AAAGCATTTC TACCACGGGA ACGACGAACC
101 AGTACAGAAA AGATAATATA AAGCGACAAT GTGTTGAGGA AAATTACGAT
151 AAAAAATTTT CTATGATTAA AAAAAAACGT CAGCAAACAT TACAAAAGTA
201 CAAGCTTAGT CTTCTGAATC CGTTGGAAAG GGCTTTTCGC CCTTTATCAT
251 ATGAAAAGTA CATGATTGGT TTAAATATGC AGTATGCATC GCATTCTCTC
301 ACTGAAGGTC ATCACTCACA TAGTTCTGCT AACGTAAAAT CACTCAATTT
351 TCCACACAGG ATACAGATAG GAGTATTAGC AAATTGTATT TTATTGGTAA
401 CTCAAGAAGG ATGTTTTCAT AGTAAATTAG TATTTGCAAC AAACAAGGGC
451 TATGTCGCAG GATTTTCATC CCTGGATAAT TTTTCTGAAG AAAATTTCTT
501 TACGGGGTTT TCCATGGCTG AATTAAATCC TATGTTGAAA TATAAGTCCG
551 AGCCAACTGA TGTGTTCAAA ACAATGAAAC TTGAGAGAAC AGTTGCCATC
601 AAGGAAGGGC CGAGCCATTA TTTTTATCAC AACGTTAATA CCAGATCTAA
651 CGTGCATACT TTTGCCATCT TTTTACAAGA CTTTTCTTCC TTAAAACTGC
701 TCAAAATTCG TCAGGTCAAA CTCAAAGAAA ATTGCCAAGT CCATGATTCT
751 CTTGTTGTTG GAGACACGTT GATTATTACC GTAAATTATC GTTGTCATTT
801 TTATGATTTG ATTCCAGAAA CCTTTCCAAA TCCCTATATC TTTTCTCCTG
851 CCAAAAGCTC AAGAAAACAC AAAAGTAGGA GTGACATTAC TTCCCTATCA
901 TTTTGTCTAC AAGAAGATGC TTTATCACCT TTAAAAAAAA CGAACACCGG
951 TGTGTTTTAT TTAGGTTACA GAAATGGTGA TTCAATGGCA ATAGTCTTCA
1001 CTAATATTAC AAACATGACA TTACAGTACT CCAAGACCAA CGGTATGACA
1051 TCAGAAAGTC GAAATCAACC TATCAGAAAT TCCCTAAAAT CTGTTGTTTC
1101 CATAAAAGCA TTGAATAATA AAGGCCTAAT ACTTATATCT GGAATGGCAG
1151 ACAAAGAGAA TGTGCAACAA CTGGTTATAG CAGATACGTT CCTGGAGGAC
1201 ATTCTGACCG AAATCCCGGT TGTATCTTTT AAAACTAAGT TCTTAAACGT
1251 GACTAAGGAT ACAGAAATAC TCGAGATATC GGATGACGGG CGCTACTTCA
1301 TTTATGGTTC AACTAGCGCT AGGGACGGAA AGGGTGATTT TGAAGTATTT
1351 TGTACCACTT TATCTGGAAA TCTTGACTAC GAAAAATCCG AAGGTGGAAA
1401 TATCACCCTC TATCCAATAG GAGGTATGAA GAACTACTGT AGGTTAGAAA
1451 ACTTCCAGTT TGAATCTATC CACCTACATT CGGCTTTCAT ACCCCCAAGG
1501 TACGTAAATC CGTTTGATGC TGTGGAACCT CTAGGTGAAG AGAGCAGTAC
1551 TTCACCTTAC GATATACCTG AAGAAGCACT TTCTCAAAAA ATATGCATCT
1601 TAATTCGGAG AGAAGATGAT CCCTACAATG GGGCCAATAT CTTCATAACA
1651 TCTGCTTTGA CCTAG
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/getSeq?map=nmap&seq=YGL176C&flankl=&flankr=&rev=>).
The length of the genomic DNA is the same as the length of the coding sequence. Thus, it does not appear that YGL176C gene contains any introns.
Protein Information
The protein encoded by YGL176C has the following primary structure of amino acids:
1 MTPNLPGFYY DRERRRYFRI SENQSISTTG TTNQYRKDNI KRQCVEENYD
51 KKFSMIKKKR QQTLQKYKLS LLNPLERAFR PLSYEKYMIG LNMQYASHSL
101 TEGHHSHSSA NVKSLNFPHR IQIGVLANCI LLVTQEGCFH SKLVFATNKG
151 YVAGFSSLDN FSEENFFTGF SMAELNPMLK YKSEPTDVFK TMKLERTVAI
201 KEGPSHYFYH NVNTRSNVHT FAIFLQDFSS LKLLKIRQVK LKENCQVHDS
251 LVVGDTLIIT VNYRCHFYDL IPETFPNPYI FSPAKSSRKH KSRSDITSLS
301 FCLQEDALSP LKKTNTGVFY LGYRNGDSMA IVFTNITNMT LQYSKTNGMT
351 SESRNQPIRN SLKSVVSIKA LNNKGLILIS GMADKENVQQ LVIADTFLED
401 ILTEIPVVSF KTKFLNVTKD TEILEISDDG RYFIYGSTSA RDGKGDFEVF
451 CTTLSGNLDY EKSEGGNITL YPIGGMKNYC RLENFQFESI HLHSAFIPPR
501 YVNPFDAVEP LGEESSTSPY DIPEEALSQK ICILIRREDD PYNGANIFIT
551 SALT*
(Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/getSeq?map=pmap&seq=YGL176C&flankl=&flankr=&rev=>).
By performing a Blastp search, I found that the protein sequence for YGL176C is similar to the itself and similar to other uncharacterized proteins in yeast strains (Fig 7). Indeed, this search is not altogether helpful.
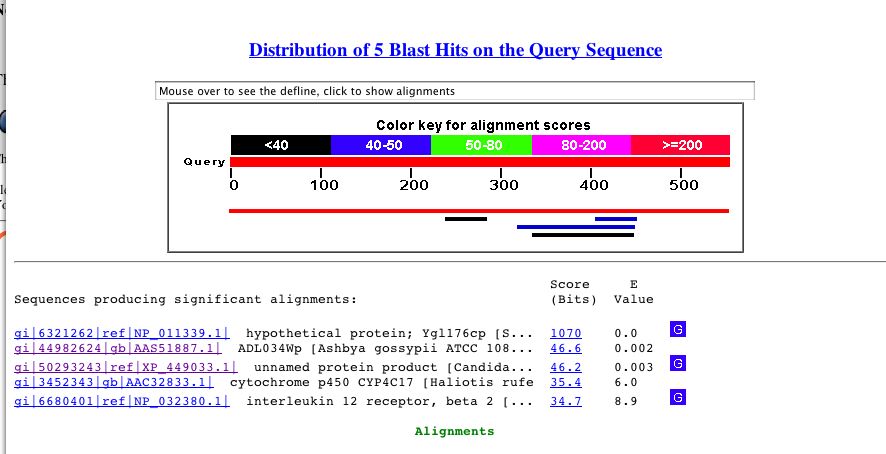
Figure 7. Blastp search results for YGL176C. Perform a search yourself by copying the protein sequence above and accessing Blastp.
I entered the amino acid sequence into PREDATOR to find out the predicted secondary structure of YGL176C.
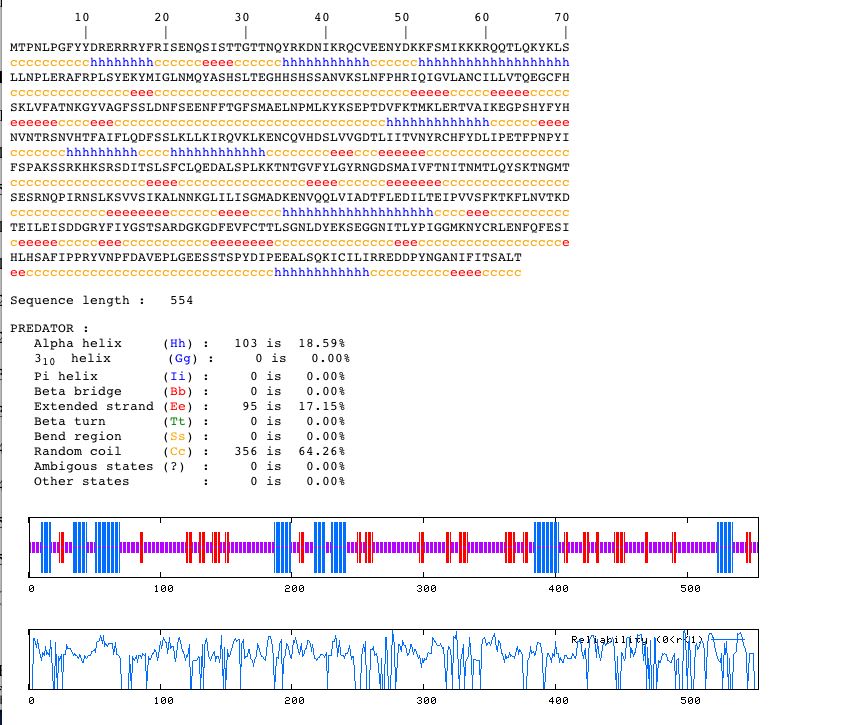
Figure 8. PREDATOR search results for YGL176C. Perform your own PREDATOR search using the amino acid sequence above (without the numbers).
From this data, it appears that the primary structure of YGL176C is composed of alpha helices, random coils, and extended strands. Molecules with a high percentage of alpha helices, often act as receptors, so perhaps this gene encodes for a receptor.
Furthermore, I entered the YGL176C amino acid sequence into the Kyte-Doolittle Hydropathy plot which assesses a protein sequence for transmembrane regions. Regions of hydrophobicity of an amino acid sequence are indicated whenever the hydropathy score is equal to or greater than 1.8. I found that the protein encoded by YGL176C appears to be a transmembrane protein, due two at least three regions of hydrophobicity. (Fig 9). Thus, the hydropathy plot data gives further support that YGL176C may be a receptor because receptors are often embedded in a membrane.
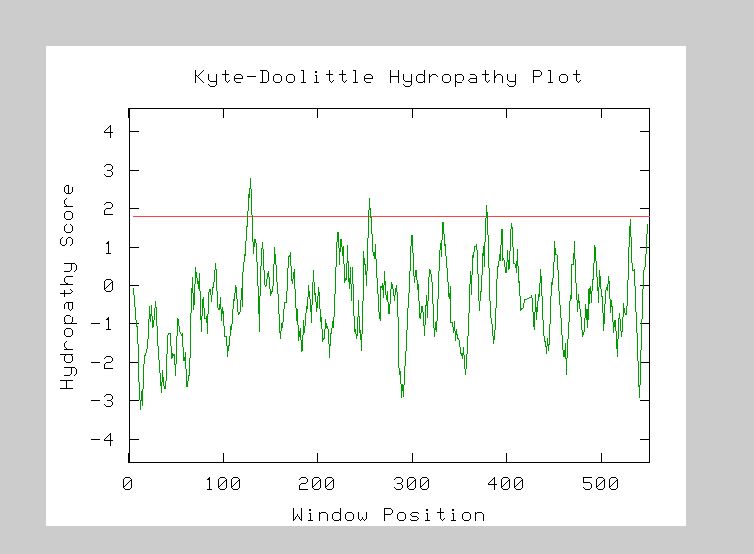
Figure 9. Kyte-Doolittle Hydropathy plot data. Perform your own KD-assessment by entering the amino acid sequence for YGL176C into the Kyte-Doolittle entry form.
Finally, I performed a protein motifs search on YGL176C on SGD website. It appears that YGL176C has a disulfide bond isomerase, DsbC, N-terminal domain (Fig 10). Thus, YGL176 may play a role in the formation of disulfide bonds.
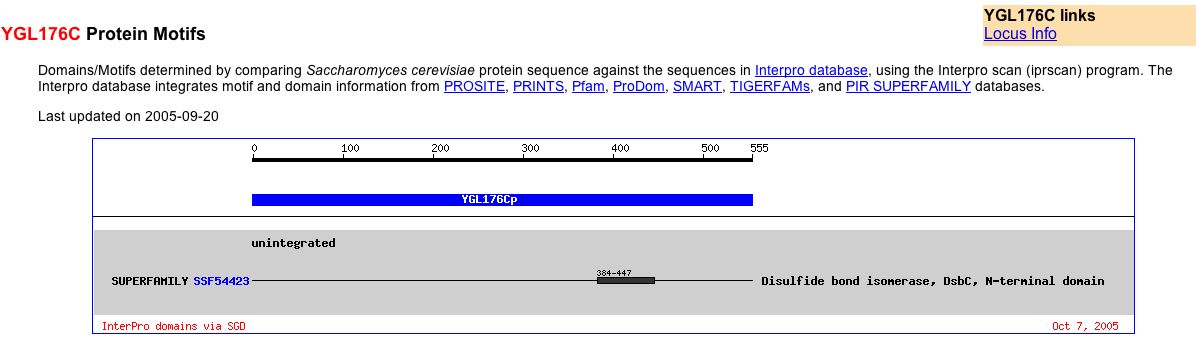
Figure 10. Protein Motif Data. (Balakrishnan R, 2005; <http://db.yeastgenome.org/cgi-bin/protein/getDomain?sgdid=S000003144>).
Mutant Phenotype:
Giaever G, et al. (2002) found that when YGL176C is systematically deleted, the mutant strain develops as viable. Thus, YGL176C is not an essential gene. Other paralogs must exist in yeast that perform the same or similar functions.
Conclusions:
The searchs above did not lead to any overwhelming evidence regarding the function or location of the protein product encoded by YGL176C. Since the protein encoded by YGL176C contains about 20% alpha-helices and because the protein appears to have transmembrane domains in the Kyte-Doolittle Hydropathy plot, I propose that it YGL176C is a receptor. The protein motif infomation does not appear to fit with my hypothesis, so perhaps alternatively, the protein product of YGL176C assists in the folding of proteins (namely disulfide bonds). Regardless, YGL176C is not an essential gene so other yeast genes share the same function or functions of YGL176C.
References:
Balakrishnan R, Christie KR, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Nash R, Oughtred R, Skrzypek M, Theesfeld CL, Binkley G, Lane C, Schroeder M, Sethuraman A, Dong S, Weng S, Miyasato S, Andrada R, Botstein D, Cherry JM. 2005. Saccharomyces Genome Database. < http://www.yeastgenome.org/>. Accessed 2005 Oct 7.
Giaever G, et al. 2002 Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-91.
Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization of in budding yeast. Nature 425: 686-691.
Rattray AJ, McGill CB, Shaefer BK, Stratern JN. 2001 May. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158: 109-122.
Roeder GS. 1997 Oct 15. Meiotic chromosomes: it takes two to tango. Genes and Dev 11: 2600-2621.