*This webpage was produced as part of an undergraduate course at Davidson College*
Paper Review
Synthetic Gene Networks That Count
Ari E. Friedland, Timothy K. Lu, Xiao Wang, David Shi, George Church, James J. Collins
Science, Vol. 324: 1199-1202. 29 May 2009. (Link)
Summary:
As easy as counting 1, 2, 3...ok, while it may not be "easy", Friedland et al. demonstrates here that synthetic gene networks can be designed to perform simple modern computing tasks such as counting. This new emerging field of Synthetic Biology pulls together techniques and ideas from various disciplines in order to construct biological systems displaying these desired consturcts. In this paper, the authors have constructed two types of biological counters in Escherichia coli that can detect and execute up to three induction events. These constructs are: 1) a riboregulated transcriptional cascade (RTC) counter and 2) a recombinase-base cascade termed the DNA invertase cascade (DIC) counter. Both des gin es utilized GFP to serve as the output indicator, which allowed the researchers to measure the relative fluorescence through flow cytometry and thus determine the level of transcription that occurred. The authors argue that modular devices like these can serve useful functions in cells which must keep track of various bimolecular processes and concentrations in order to effectively regulate metabolism and growth. They suggest that circuits similar to their constructs could be used for biosensing, bioremediation or medical purposes. While their system is not completely fool proof in that some noise or leakage still occurred, for the most part their mathematical models were able to accurately predict the experimental resutls--thus confirming their designed constructs.
Methods:
The methodology used in this paper mainly involved the creating of the two counting constructs. Both types of counters were designed in plasmids that were then later transformed into Escherichia coli cells. All other techniques were relatively simple molecular biology techniques.
Riboregulated Transcriptional Cascade (RTC): This modular design consisted of a two-counter and three-counter system that was inducible by the researchers in order to promote transcription of a desired gene, in this case GFP. The two-counter and three-counter systems differed only in that the three-counter construct had an additional riboregulated step, the production of T3 RNAP which would then induce the T3 promoter to translate GFP. The RTC constructs consisted of: a promoter, a cis repressor (cr) segment, the Ribosomal binding site (RBS), and a gene. The cr sequence was placed in between the promoter and RBS site. Upon induction a stem-loop secondary structure would form preventing binding of the 30s ribosomal subunit thus inhibiting translation. However, if a second inducible activation occurred, the activation of PBAD by arabinase, then taRNA would be transcribed and would bind to cr inhibiting its binding of RBS. This inhibition of cr would then allow for successful promotion of downstream fragments, resulting in GFP production. These RTC counters serve as models for time dependent cellular events such as cell division because of the fast activation of the transcriptional and translational regulatory elements used in these devices.
DNA Invertase Cascade (DIC):This design consisted of a single inducer and multiple inducer system that strung together modular DNA-based counting segments. These units used natural recombinases Cre and flpe which could lead to the inversion of DNA between two recognition sites loxP and flpe-. Along with other fragments such as RBS and ssrA, which causes the rapid degradation of protein, these recombinase segments were linked in a chain along with their desired promoters creating what was to be called the Single Invertase Memory Module (SIMM). These SIMMs were placed in a particular order so that with each pulse of the inducer they would activate the promoter causing a flip in the DNA segment and thus leading to the production of the final segment encoding GFP. This system of constructs operates on a time scale of hours, whereas the RTC constructs are designed for fast activation, the DIC design would allow the researchers to study sequential events in areas such as developmental biology and/or gene cascades.
Figure 1: The RTC two-counter and RTC three-counter construct designs and results.
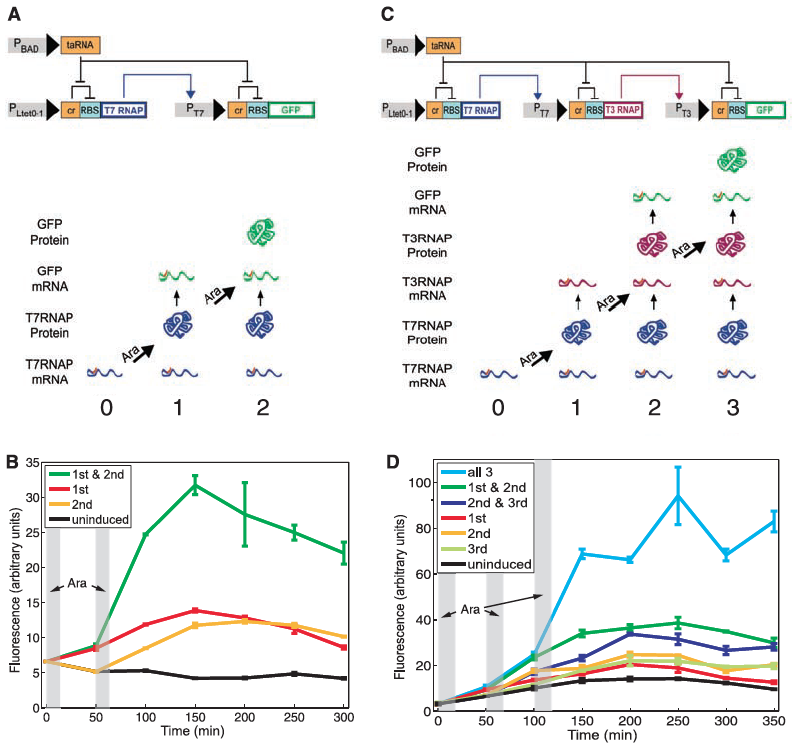
A) Diagramed here is the RTC two-counter construct. The top portion of part A shows the construct itself and and the lower half illustrates the different outcomes when subjected to the pulse. For the construct to work properly the Ltet0-1 promoter is induced by a pulse which drives transcription of T7 RNA polymerase. This then binds to the the T7 promoter and finally transcribes GFP. The riboregulation of this construct is determined by the activation of taRNA induced by arabinose pulses which cases the removal of the cr inhibitor of RBS to allow for the correct transcription of GFP. Arrows indicate the direction of activation while flat-tipped arrows indicate inhibition of signal transduction. As for the lower illustration, this is just a simplified representation of the effects of each pulse. Since the cell starts with T7 RNAP mRNA already present at time point 0, the first pulse allows for the T7 RNAP to bind to the T7 promoter thus initiating GFP (time point 1). The second pulse allows allows for the more translation of GFP mRNA leading to high fluorescence.
B) This panel simply shows the mean fluorescence of the RTC two-counter system. Fluorescence is measured over time by a flow cytometer and plotted as four separate lines: uninduced, 1st pulse, 2nd pulse, 1st & 2nd pulse. The gray columns indicate when each arabinose pulse was applied. Interestingly only constructs that received both the 1st & 2nd pulse were fully activated. Despite seeing some leakage in fluorescence, only the commination of both pulses resulted in optimal activation, and all activation regardless of the pulse treatment occurred at the time point of the second pulse. This figure simply showed that the RTC two-counter construct was working properly despite seeing a little noise.
C) Diagramed here is the RTC three-counter construct. Again the top portio shows the construct design and the lower half the outcomes when subject to the various pulses. As noted in the methods section, this construct only differs in the additional riboregulation step. This part of figure 1 shares the same purpose as did part A--to illustrate their designed construct.
D) Again, as in part B, this panel shows the experimental output for the RTC three-counter construct. Similar patterns of induction again were verified as was in part B.
Figure 2: Modeling predictions and RTC three-counter experimental characterization.
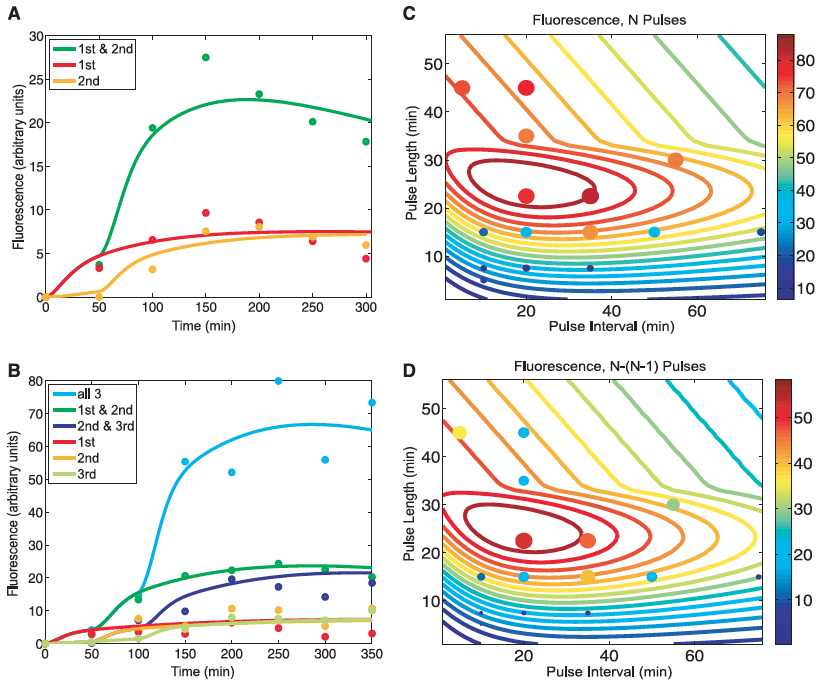
A) This panel compares the experimental results for the RTC two-counter construct with those predicted by mathematical modeling. Lines represent the predicted fluorescence levels for cells receiving the different combination of pulses. Dots indicated the experimental results of the construct under the same combination of pulses. This figure serves as a proof of concept as indicated by the close matching of the lines and dots for each condition.
B) Same as in A but this time for the RTC three-counter construct. Again, a close match is seen between the experimental and predicted values. This again validates their results.
C) This panel shows the outcomes of the mathematical model predictions across a range of pulse lengths and time intervals for the RTC three-counter construct (designated at "N Pulses" above the figure). The sprial contour lines show the predicted fluorescence and dots illustrate the experimental results. Color and size of the circles indicate the expression levels for these experimental results. This figure suggests that max expression occurs with pulse lengths of ~20 to 30 minutes and interval of 10 to 40 minutes.
D) Here the absolute difference in fluorescence between three pulses (N) and two pulses (N-1) is shown. This figure is similar to part C in that lines again are predicted values and dots are experimental. This figure confirmed the optimal counting conditions observed in part C. Also, it suggests that since the model predictions were consistent across a wide range, that the RTC three-counter construct has a robust counting behavior over a fairly large temporal region
Figure 3: The single-inducer DIC three-counter construct design and results.
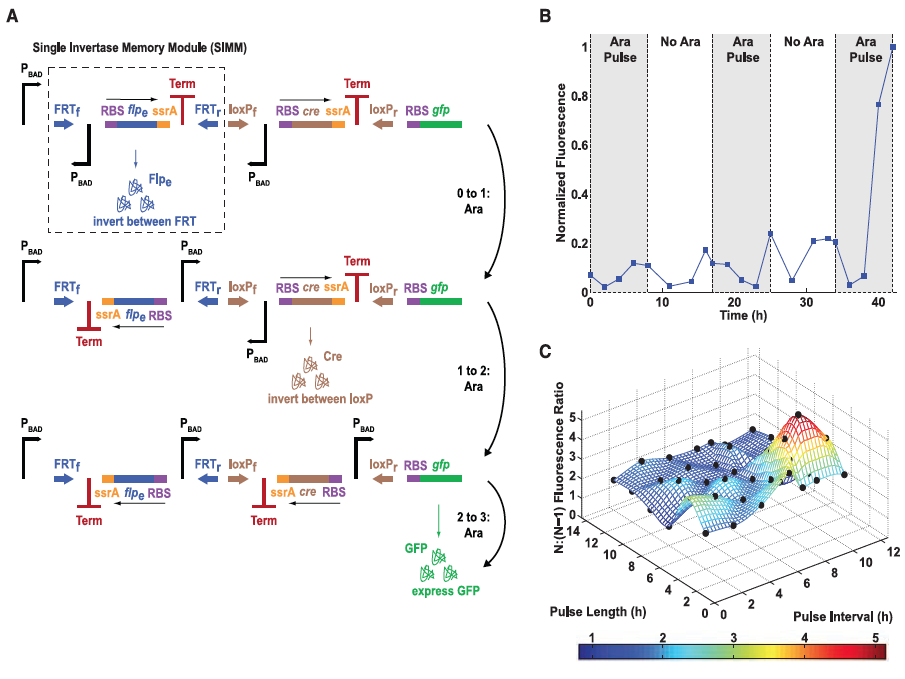
A) Here the authors illustrate their second type of design, the single-inducer DIC three-counter construct. This construct is comprised of a number of Single Invertae Memory Module (SIMM), one of which is outlined by the dashed box. GFP is used again as the reporter gene so that successful expression can be measured. Each pulse causes signal transduction to occur along the pathways outlined by the arrows to the right of the figure which results in flipping of the SIMM segment and the transcription of the next segment. The third and final pulse translates and transcribes GFP, which is then measured by flow cytometry.
B) Mean fluorescence is shown again for the experimental data as was in Figure 1 but this time for the singe-inducer DIC three-counter construct. Some small activation occurred after the second pulse but full activation was not observed until the final third pulse was applied. This figure confirmed their construct because by design full activation should not occur unless in the presence of all three pulses of arabinose (each indicated by the shaded regions in the graph).
C) This last part compares GFP ratios between three pulses of arabinose (N) versus two pulses of arabinose (N-1) for the single-inducer DIC three-counter construct for experimental and predicted models. Experimental results are illustrated as black dots. As shown, the predicted values matched very closely to those determined experimentally. This demonstrated that the single-inducer DIC three-counter construct was able to effectively count pulses of varying length and intensity in the range of 2 to 12 hours.
Figure 4: The multiple-inducer DIC three-counter construct design and results.
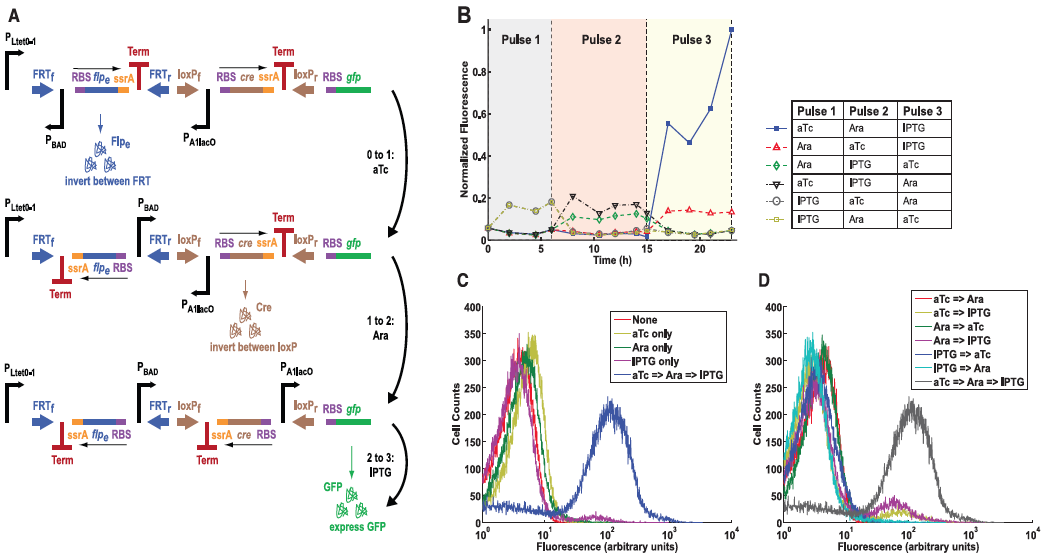
A) This panel diagrams the authors improvement from the construct in Figure 3, a multiple-inducer DIC three-counter construct. This new design uses three different promoters instead of simply just P BAD. Here they show the same ideas as in Fig. 3A, except the first pulse is anhydrotetracycline (aTc), the second pulse as arabinose, and the third being IPTG. All three pulses activate their respective promoters and again lead to GFP production.
B) This graph shows the mean fluorescence determined by flow cytometry. Each combination of pulses was tested as indicated by the legend to the right of the graph. Again, only high amounts of GFP was observed when all three pulses were performed. This results suggests that the circuit can be programmed to respond only to a desired sequence of events.
C) This panel simply compares constructs that received only one type of pulse versus all three. Again fluorescence was measured by flow cytometry and plotted along the graph. Again, only those receiving all three pulses resulted in high fluorescence, all others demonstrated low fluorescence.
D) Finally, this panel is similar to part C but this time shows different combinations of two pulses versus constructs receiving all three. Again, only cells that received all three pulses showed the correct levels of fluorescence. This graph simply re-confirmed their conclusions from previous figures.
Conclusion & Critique:
I thought this paper provided a very interesting novel application of synthetic biology. They provided adequate information to support their claims and back this with more explanation in the text of the paper. However, I would have like to see why a N-(N-1) comparison was prefomed. This was not well explained in the figure legends nor the supporting text. All in all I thought the authors did a very good job at explaining their concepts. They performed many of their expriments multiple times which leads me to believe their data even more so. Also they did a very good job explaining exactly their design of each construct despite the combination od DIC constructs being very complicated. Overall I like this paper very much and thought that it provided many convincing arguments about the application of designed synthetic devices such as these.
References:
Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic Gene Networks that Count. Science. 2009 May 29;324: 1199–1202.
William's Home Page
Genomics Page
Biology Home Page
Email: William Green for questions or comments.
© Copyright 2011 Department of Biology, Davidson College, Davidson, NC 28035