This web page was produced as an assignment for an undergraduate course at Davidson College.
Summary:
Cis-regulatory modules (CRMs) are pieces of DNA that provide transcription factors a place to bind, so that they can control the translation of nearby genes. CRMs provide a selective advantage for coordinately expressing genes involved in a biologically pathway all at the same time. Two genes, TMEM216 and TMEM138, which are nonparalogous genes, were shown to separately cause Joubert syndrome. They discovered that the deleterious mutations found in the TMEM216 gene on chromosome 11 only accounted for half of the ten Joubert syndrome families being studied, even though the two groups were phenotypically indistinguishable. To identify the alternative gene, researchers resequenced the exogenic and promoter elements in the 17-Mb candidate region around TMEM216. They found four missense mutations and one splicing homozygous deleterious mutation in the TMEM138 to account for the remaining families. This was a strange case because the two genes represented two distinct protein families, did not demonstrate any sequence homogeny, or share any functional domains; the only commonality was that they were transmembrane proteins.
Since the genes were aligned directly next to each other, due to an ancient chromosomal rearrangement at the amphibian to reptile evolutionary transition, the researchers tested if the genes were part of a CRM and in turn, coregulated and cofunctioning. They did this by testing tissue-expression patterns in humans and mRNA levels in zebra fish versus mice. They found tight coexpression across major tissues and tight coordinated mRNA expression levels in mice compared to zebrafish, whose TMEM216 and TMEM138 are not directly adjacent. They also noted that both genes were needed for ciliogenesis and could be found tagging separate vesicles that localized to the base of cilia and contained different sets of cilia-targeted proteins. They then used siRNA to knockdown each protien in turn to test if TMEM138 and TMEM216 were required to move the vesicles containing the other. They found that TMEM216 was required for the correct movement of the TMEM138 vesicles. They found retention of the coordinated movement of TMEM216 and TMEM138 vesicles in zebrafish, which suggests that the coordinated localization preceded coordinated gene regulation. However, the two genes in zebrafish caused different phenotypes, unlike in humans, which would suggest that gene function changed to be more similar with the chromosome rearrangement.
Figures:

Figure 1:
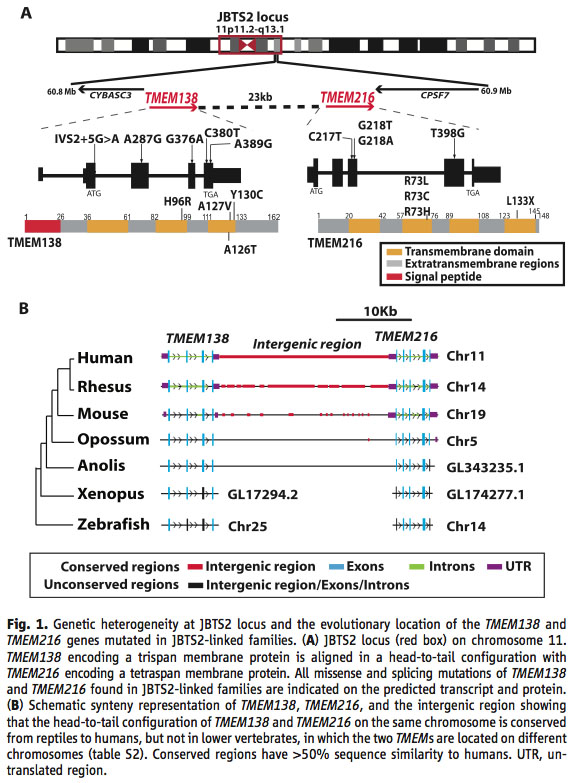
A) The first line is showing the evolutionary location of theJBTS2 locus, which represents the genes that can cause Joubert syndrome on chromosome 11. The second line shows that the genes are lined up on the chromosome in a head-to-tail configuration with a 23 kb pair noncoding intergenic region between them. The third line shows all of the identified missense and splicing mutations found in the two genes that caused a disease phenotype in the studied families. The fourth line is a diagram of the final protein, showing transmembrane domains in yellow, extratransmembrane regions in gray and signal peptides, which direct the transport of the protein in red.
B) Shows a synteny representation of the two genes and their intergenic region over a group of evolutionary divergent species. The important thing to note here is the conservation of linkage and an equivalently long intergenic region across species after the divergence of amphibians and reptiles.
I thought panel A in figure 1 was a waste of space because that was not what the paper was trying to show. It was showing information that was easily explainable in words, information was just a background to the rest of the paper, didn’t need to be proved to the reader, and is more succinctly shown in panel B.
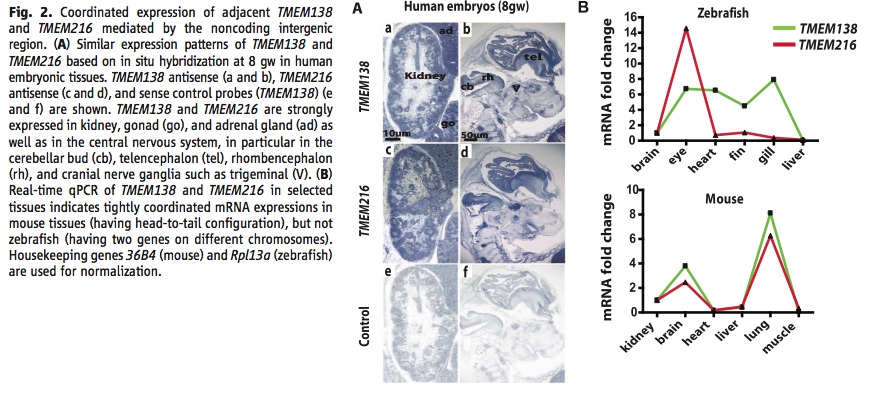
Figure 2:
A) This panel is showing the similar expression patterns of the TMEM138 and TMEM216 proteins in human embryonic tissues detected by in situ hybridization. TMEM138 expression in shown in a and b, TMEM216 in c and d, and a control in e and f using sense control probes. The tissues representing high coexpression are kidney, gonad (go), adrenal gland (ad), cerebellar bud (cb), telencephalon (tel), rhombencephalon (rh), and trigeminal (V).
B) This is a graph of real-time qPCR done on both TMEM138 and TMEM216 in selected tissues in mouse and zebrafish. They used species from both sides of the evolutionary chromosomal rearrangement to see if the linkage of the genes affected expression. They found tight coordination in mouse tissues over zebrafish tissues, which give support to both genes being involved in a CRM.
I had a few problems with figure 2 B. First of all the y-axis is measuring a fold change, but the figure never tells you what initial values they are measuring against to create a fold change. Secondly, they do not have error bars on their data points, so the reader has no idea if they ran multiple trials or how representative the data is of the typical trend. It is good that they included the genes used for normalization because those can affect the qPCR values and would be helpful for replication by a reader.
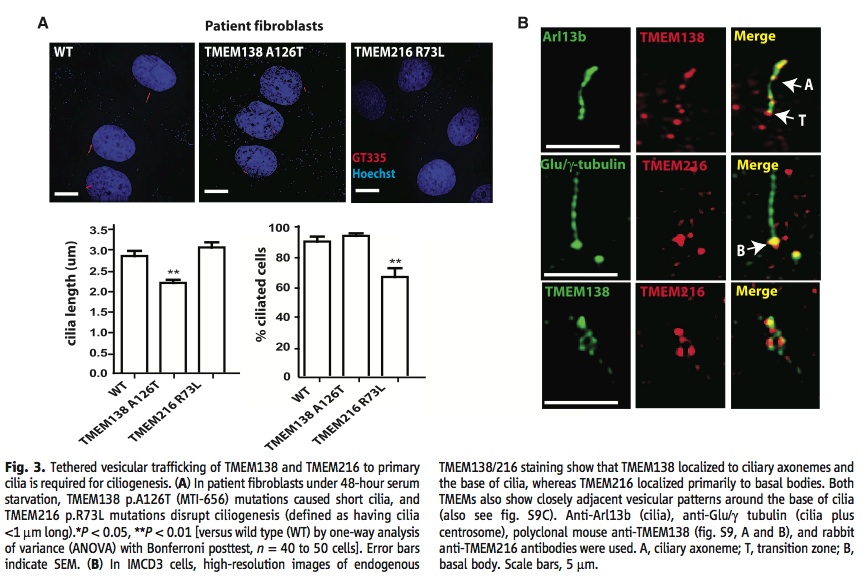
Figure 3:
A) The left graph is showing that mutations in the TMEM138 gene cause shorter cilia than wild type or a mutated TMEM216 gene. The right graph is showing that there are significantly less total cilia being produced when there is a mutation in the TMEM216 gene compared to wild type and a mutated TMEM138 gene.
B) These images are showing where the TMEM216 and TMEM138 proteins localize in IMCD3 cells. TMEM216 localized primarily to ciliary axonemes and the base of cilia, while TMEM216 localized primarily to basal bodies. Anti-Arl13b was used to show the location of cilia and anti-Glu/γ-tubulin fluoresced the cilia and centrosome. In the image A represents ciliary axoneme, T, transition zone, and B, basal body.
The images of the patient fibroblasts were not helpful in understanding the paper and were not very compelling either. They were no significant and noticeable differences between mutations, which would be the reason for including the images.
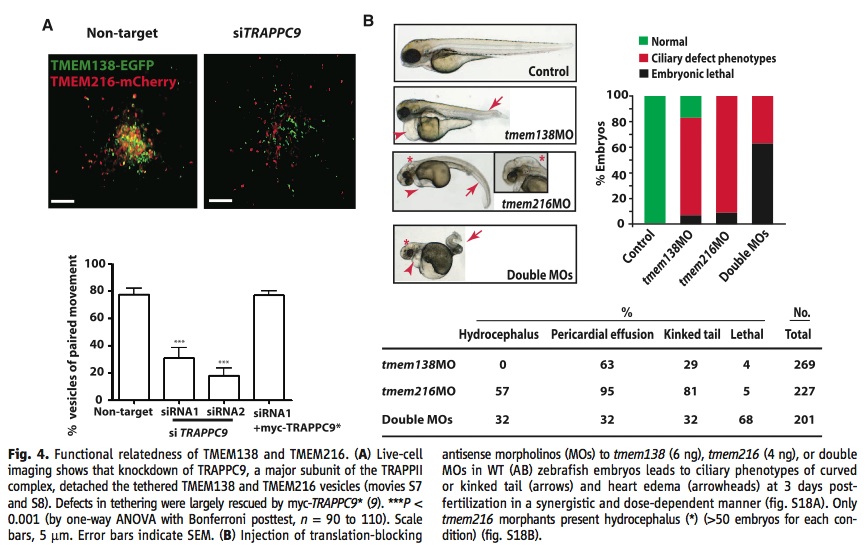
Figure 4:
A) These images and graphs show that TRAPPC9, which is an integral component of the TRAPPII complex, is necessary to keep the TMEM216 and TMEM138 vesicles tethered to each other. The non-target RNA image and bar on the graph show a large amount of yellow dots and a high number of vesicles with paired movements, while addition of siTRAPP9 significantly decreased the number of yellow dots and the number of vesicles with paired movements.
B) This panel is showing that in zebrafish knocking out one of the proteins does not produce the same phenotype as knocking out the other one would. They knocked out each protein separately and then together by injecting antisense morpholinos to stop translation in zebrafish embryos.
I didn’t understand why they included A in figure 4. It did back up their claim that TRAPPII was integral for the paired movement of vesicles, but the main point of the paper was to identify main functions and relationships between TMEM216 and TMEM138. Since TRAPPII was not the study genes I don’t think space needs to be sacrificed in the paper to add images about it and the figures might have been better placed in the supplemental material.
Conclusion:
I thought it was smart of the researchers to keep their search focused on the region around the TMEM216 gene initially because evolution often mutates DNA to put proteins used in similar pathways next to each other. It was also a good idea to try and identify if the mutations they found in the new gene TMEM138, were VNTRs or SNPs by trying to find them in other people, since that gave them more evidence for TMEM138 being the correct second gene. The comparisons between species before and after the rearrangement gave clear results that showed that there were significant differences between the two groups. Overall, I feel like the authors gave the reader sufficient evidence to prove that their experiements "suggest that nonparalogous genes not only can be chromosomally rearranged into a functional gene cluster during vertebrate evolution, but also can be assembled into a new CRM by evolving regulatory elements, which correlate with their coordinated expression" (Lee et al., 2012).
References:
Jeong Ho Lee, et al. 2012. Evolutionarily Assembled cis-Regulatory Module at a Human Ciliopathy Locus. Science, 335(6071): 966-969.
Scientific Article on Evolutionarily Assembeled cis-Regulatory Module
Genomics Page
Biology Home Page
Email Questions or Comments to ermcguire@davidson.edu.
© Copyright 2011 Department of Biology, Davidson College, Davidson, NC 28035