Chris Peek
In Situ Evolutionary Rate Measurements Show Ecological Success of Recently Emerged Bacterial Hybrids
Vincent J. Denef and Jillian F. Banfield
Introduction
Analyzing how a population evolves in real time is a crucial question for many genomicists that would provide valuable insight into the mechanisms of natural selection. However, time constraints limit the feasibility of this question to species that produce offspring at a rapid rate. Accordingly, bacterial communities provide an excellent model for addressing this question. Additionally, analyzing the evolution of bacterial communities is of particular interest because of the crucial roles they play in a myriad of fluctuating ecological processes. Many groups have analyzed bacterial evolution by adding selection pressures artificially in lab, or by allowing natural samples to grow in a laboratory setting. However, such experiments fail to truly capture the evolution of a species in its natural habitat. Denef and Banfield have overcome these limitations by directly sequencing the genome of a bacterial community over a period of 9 years. The group chose a feasible experimental location and system to work with – one characterized as a discrete, well-defined, relatively homogenous community. Additionally, the researchers ensured both the reproducibility of their community of choice and the limited input of outside microbial species. The bacterial community in the acid mine drainage (AMD) in the Richmond Mine in California meets the criteria. Denef and Banfield chose to focus their efforts on characterizing the evolution of the dominant primary producer in the AMD biofilm, Leptospirillum group II. High-throughput sequencing of AMD biofilms over an extended period of times allowed Denef and Banfield to accurately calculate an in situ mutation rate for this bacteria, map the major recombination events that led to various subpopulations (types) of Leptospirillum group II, and support the hypothesis of rapid positive selection favoring Leptospirillum group II hybrids. Below, I will summarize the results in detail. (DeLong, 2012; Denef and Banfield, 2012).
Glossary
- AMD = Acid Mine Drainage site
- Leptospirillum group II – Species of chemolithoautrophic bacteria found in the AMD biofilm; includes types I, III, IV, IVa, V, and VI
Notable Previous Work
- Reconstruction of Leptospirillum group II types I and VI genomes
- Evidence for recombinant genotypes between Leptospirillum group II types I and VI
Results
Figure 1
This figure addresses the question how bacterial community diversity changes at various sampling sites over time and with respect to environmental change. Both Illumina sequencing and FISH analysis are used to generate these data.
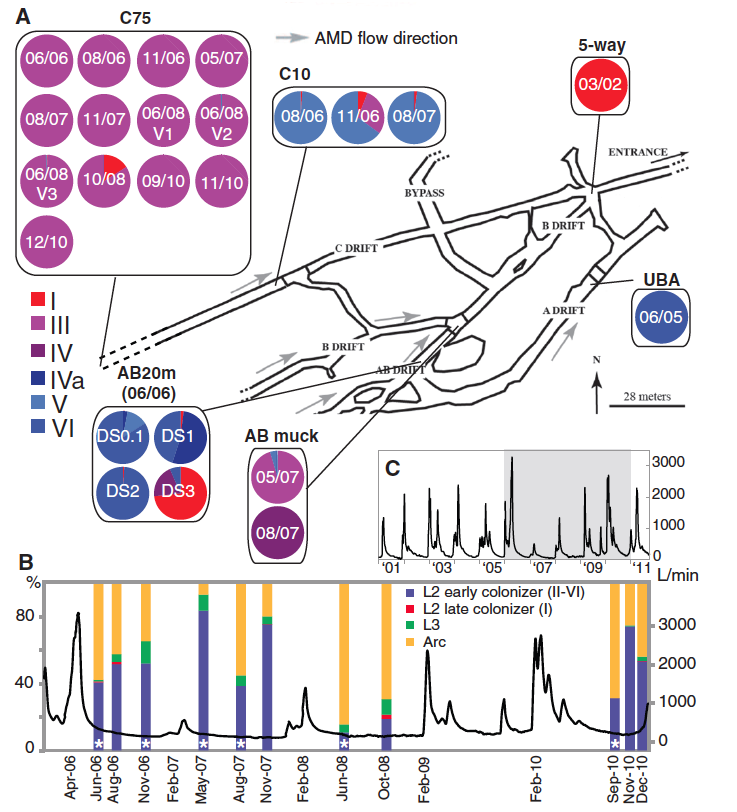
(Figure 1 Denef and Banfield, 2012; permission pending)
Panel A: This panel provides a schematic of the AMD system that indicates the sampling sites and flow direction. The results of high-throughput sequencing of biofilm samples are displayed in the pie charts for each sampling site. These charts show the proportion of each Leptospirillum group II type at each sample on the indicated sampling data. These sequencing reads for the recombinant genotypes (III, IV, IVa, V, and VI) were recruited to the parental type I and VI genomes. Quality control was employed by limiting alignment reads to the highest similarity. Coverage from the Illumina sequencing ranged from 1.7 to 8 million reads per sample. Site C75 contains primarily type III Leptospirillum. By comparing the high frequency SNPs fix in the type I-like and type VI-like segments of the type III genome to the type I and type VI parent genomes, the researchers were able to calculate a mutation rate of 1.4 x 10-9 substitutions per nucleotide per generation. Denef and Banfield also checked for potential spatial variation in the population by sampling three locations ~1m apart at the C75 site (06/08 V1, V2, and V3). Because they found little variation between these sites, the researchers concluded that they were not biasing their results by excluding potential spatial variation. The difference of bacterial community composition along the same flow path (C75 vs. C10) suggests positive adaptive selection for different environments. The fact that the type III Leptospirillum found at C10 was a different genome than that found at the C75 location supports this hypothesis. Furthermore, this diversity indicates that adaptive fitness rather than lack of diversity might explain the dominance of single genotypes in a biofilm.
Panel B: These data show bacterial community population composition over time aligned with the AMD flow rate for the C75 sample site. Fluorescence in situ hybridization (FISH) was used to determine the presence and abundance of each indicated species from biofilm samples. One can see that high abundance of type III Leptospirillum (asterisks) and how high flow rates can alter bacterial community composition.
Panel C: This panel shows the AMD flow data on a larger time scale (2001-2011). The shaded box indicates the region shown in panel B.
Figure 2
This figure attempts to reconstruct the evolutionary history of Leptospirillum group II by mapping major homologous recombination events via proteomics-inferred genotyping (PIGT) and Illumina sequencing. Ultimately, this figure supports the hypothesis that major recombination events have recently driven Leptospirillum evolution.
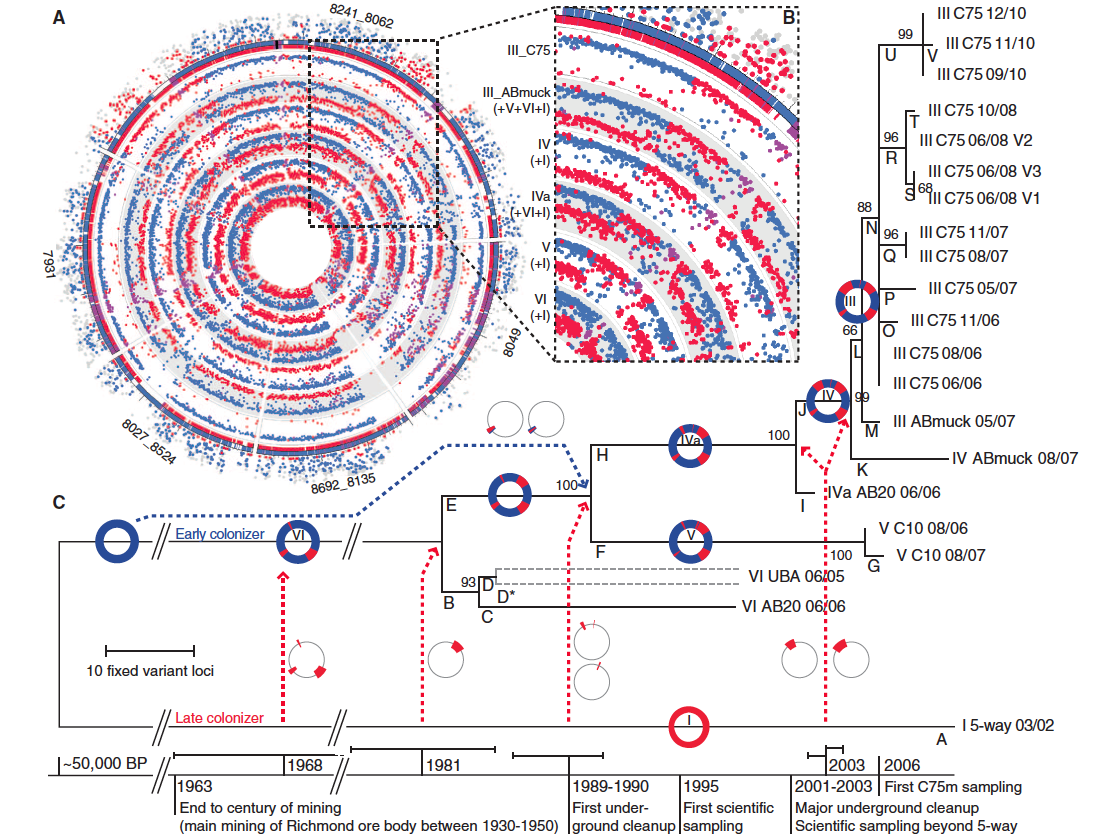
(Figure 2 Denef and Banfield, 2012; permission pending)
Panel A: The outer circle of dots indicates the unique peptides to type I (red) and type VI (blue) reference genomes present in the reconstructed type III C75 population (purple indicates the presence of both type I and type VI). The inner circles depict the Illumina high-throughput sequencing data of the recombinants III, IV, IVa, V, and VI. One can see the major recombination events that took place by comparing each recombinant genotype to the parental genotypes (innermost circles).
Panel B: This inset highlights identical recombination points in Leptospirillum group II types III, IV, IVa, and V.
Panel C: This panel shows a phylogenetic tree based on variant loci. A timeline along with inferred recombination events from panels A and B is shown below, suggesting a mechanism for how each Leptospirillum group II genotype arose. While the timeline also denotes human events, the researchers are not able to determine whether human natural changes in the AMD environment drive the observed mutations. This timeline was generated using the substitution rate previously described. Statistical analysis of populations in this tree provides evidence of positive selection for type specific mutations.
Figure 3
This figure addresses what types of mutations became fixed in the various Leptospirillum types, and ultimately supports the hypothesis that positive selection, and not random mutation, is the driving force for the observed mutations. This conclusion is based on an overrepresentation of fixed mutations occurring in global regulators, signal transduction genes, and transcriptional regulation genes, mutations in which have been shown to be important driving factors for ecological divergence. These data were generated by SNP analysis of high-frequency variants. Mutations were then functionally categorized.
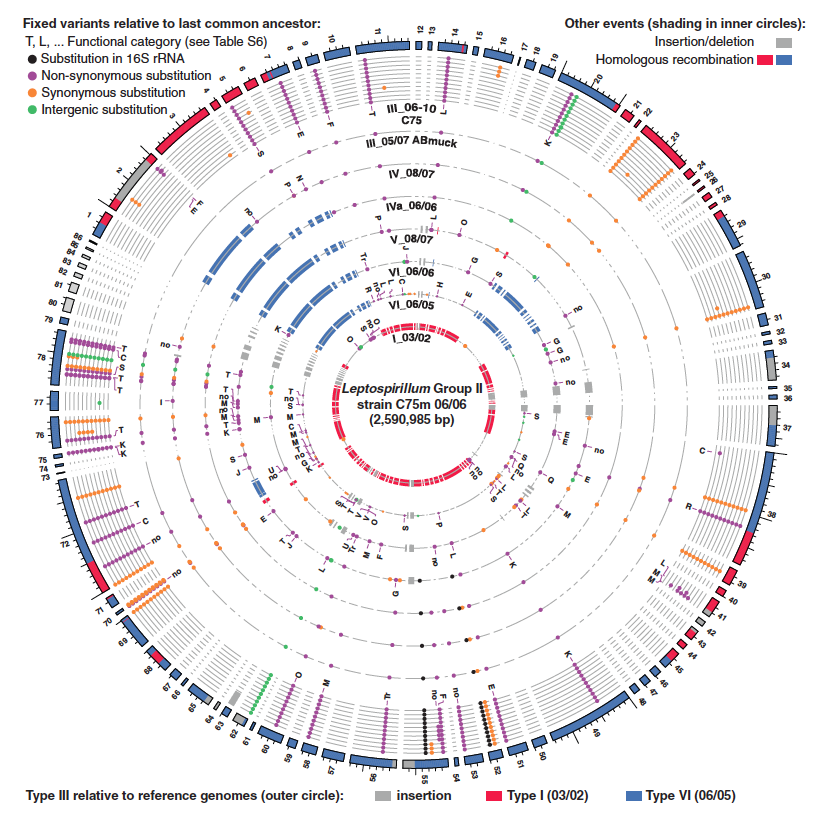
(Figure 3 Denef and Banfield, 2012; permission pending)
This plot shows high-frequency SNPs of various Leptospirillum type genomes compared to the C75 June 2006 type III sample. One notes the frequency of mutations occurring in signal transduction genes (T) and transcriptional regulation genes (K). Interestingly, one can see the accumulation of mutations in the same type III at different locations and time. The red and blue on the outer circle indicate type I- and type VI-like regions within the type III C75 reference genome. The red and blue shown in the inner circles depict homologous recombination events. Dots indicate fixed variants of the indicated category.
Summary and Evaluation
In an exhaustive study, Denef and Banfield were able to monitor the evolution of Leptospirillum group II in real time. In doing so, they were able to calculate the in situ mutation rate and reconstruct an evolutionary history of the Leptospirillum group II types I, III, IV, IVa, V, and VI by mapping major homologous recombination events. The group was also able to reconstruct the Leptospirillum group II type III genome de novo. Even more, Denef and Banfield provide several lines of evidence supporting the hypothesis that rapid positive selection has acted in upon Leptospirillum group II to fix advantage SNPs into the various populations. Such evidence suggests that individual species fitness rather than lower biodiversity plays a role in explaining the dominance of a single genotype in a bacterial community. These findings provide novel insight into the mechanisms driving evolution, and provide a deeper understanding of how bacterial communities adapt to changing environments.
Almost as daunting as the original challenge of tracking evolution of a bacterial population in situ is the task of presenting the massive amount data from this study in digestible, user-friendly manner. Taking into account the journal limitations, I believe that Denef and Banfield did a very nice job of this. By primarily focusing on the C75 site, the authors tell one of what could have likely been many compelling stories. In doing so, they avoid overwhelming the reader, which is particularly impressive given the novelty of their research. The data interpretation was sound and the authors acknowledged the limitations of their study. The paper was well organized and flowed from one section to the other in an easy to read manner. However, the authors could have annotated some figures in more detail (specifically the functional categories in figure 3), and provided a list of future directions they might wish to pursue after these findings. The repeated references to supplemental material also slightly encumbered the reading of this paper. Even taking these criticisms into account, Denef and Banfield provide an exceptional research study and present the data in a digestible fashion.
References
Genomics Page
Biology Home Page
©
Copyright 2013 Department of Biology, Davidson College, Davidson, NC 28035