This web page was produced as an assignment for an undergraduate course at Davidson College.
Autism spectrum disorders (ASDs; also known singularly as autism), are neurodevelopmental disabilities characterized by impairments of normal social interaction behaviors, impairments of verbal and nonverbal communication abilities, repetitive behaviors and potentially other cognitive of physical deficits. ASDs vary in both symptom type and symptom severity, and include diseases such as autistic disorder, Rett syndrome, and the condition formerly known as Asperger’s syndrome, among others. Some individuals with autism may also show exceptional abilities in a specific subject such as math or music [1]. The genetic correlates of autism are complex and still being explored.
A PBS feature describing a specific example of some of
the behavioral aspects of the condition can be viewed here.
More information regarding prevalence and anticipated
causes of the disease can be found here.
How were gene variants associated with autism discovered?
A recent
study by Matsunami and colleagues (2013, [2]) provides one example
of how progress is made in identifying possible genetic bases of
conditions such as autism. This study followed up on previous studies that
suggested that certain copy number variants (CNVs; differences in the
number of adjacent repeats of a given nucleotide sequence) may be largely
responsible for certain cases of autism and attempted to identify such
specific CNVs that may hold particular disease relevance. Researchers
first identified CNVs occurring in families found to be at high-risk for
ASDs, then compared the CNVs identified, along with CNVs identified from
other publications, to those in a larger sample set of child ASD cases as
well as controls (children without ASDs).
This team needed to discern the genotype of each of the
candidate CNVs within each of the 2,185 child ASD cases and 5,801 control
cases inspected in order to discern whether any of their candidate CNVs –
including 185 CNVs reported previously in scientific literature to be
potentially linked to ASDs – were more frequently associated with ASD
opposed to controls. To efficiently obtain such a massive amount of data,
researchers used a custom microarray
with probes within and flanking each of the CNV candidate regions.
Comparisons across genotypes highlighted 15 CNVs highly associated with
ASDs that researchers predicted would be useful in genetic screens for
ASDs. Further measures also probed for 2,799 single nucleotide variants
(SNVs; variations in single nucleotides that have only been identified in
a small number of individuals) previously reported in the literature to be
potentially linked to ASDs. These analyses revealed 25 CNVs not all from
the candidate pool of CNVs, expanding the body of known gene variants
linked to ASDs. The authors address that further studies will likely be
necessary link specific genetic variations to subtypes within the body of
ASDs.

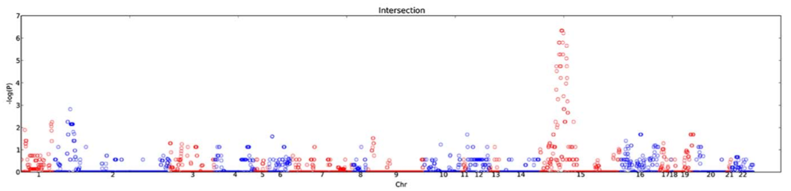
Figure 1. Manhattan plot of the CNVs analyzed at select chromosomal loci using custom microarrays. The -log (P) value reflects an odds ratio of a given CNV being identified in the ASD group compared to the normal group. CNVs with an odds ration of greater than 2 were considered clinically relevant to ASDs. Chromosomes are depicted in an alternating red and blue pattern by number for visualization. Figure adapted from Matsunami, et al. (2013). Permission pending.
How were these findings interpreted and reported by the popular
press?
Findings such as those made by Matsunami and colleagues
(2013) are regularly translated form the language of academia to that of
the general public by organizations within the popular press. These
translations vary in their framing of the research process and their
presentation of the content of the research itself. Authors of primary
literature in the sciences are relatively unrestricted in their use of
technical jargon, especially throughout the descriptions of their methods,
as persons fairly educated in the article’s focus typically read these
publications. To make information in this more technical form both
informative and useful to the public, the popular press has the
potentially-challenging role of translating these scientific findings into
more approachable language, often in the face of further challenges such
as space limitations and audience interest [3]. One admirable example of
popular press reporting of the particular study by Matsunami and
colleagues just discussed can be found in a recent
article in TIME, entitled "New Gene Variants Linked to
Autism." This article begins with an concise summary of the findings made
by the researchers, stating,
"In one of the largest-ever studies of genetics and autism, researchers have identified 24 new gene variants associated with autism spectrum disorders (ASD). The work also confirms that 31 variants previously linked to the developmental disorder may serve as useful genetic markers for identifying those with the condition."
This summary identifies specific aspects of the results without using
excess jargon, and manages to strongly suggest the potential clinical
importance of the discovery very concisely. The focus then moves directly
to the relevant context of the study, discussing the general clinical
relevance of these discoveries. Links included provide some more specific
elaboration on why such improvements in diagnostic methods are of
importance for management of the disease:
"Understanding autism’s genetic roots is a priority, researchers say, since it may lead to earlier diagnosis and behavioral intervention, which can improve patient outcomes. 'Oftentimes findings like this get published in academic journals, but they don’t get translated into clinical use,' says Chuck Hensel, an author on the new research study, published in PLoS ONE, who is the senior manager of research at the genetic diagnostics company Lineagen. 'Our goal,' Hensel says, 'is to try to get these markers into the clinic.'"
The quote from Hensel embodies not just the goals of this study, but an
emerging goal of this manner of research in general, firmly establishing
the practical context of the study. The article then transitions into
introducing the research process itself, and the interaction of the
organizations involved in this project, beginning a narrative to bring the
reader into the process of research as it was organized and carried out:
"Hensel teamed with researchers at the University of Utah and the Children’s Hospital of Philadelphia and devised a two-pronged approach for hunting down genetic markers of autism. First, the researchers chose 55 people living with autism, all from families with many members diagnosed with ASDs. The scientists then sequenced the genomes of these subjects, and compared the genetic profiles to those from a reference population, using the Utah Genetics Reference Project. That allowed them to find regions where the autistic individuals differed from people without the disorder, and led to 153 gene variants, or genetic red flags for the condition."
Jargon is practically avoided entirely as the project plan is laid out,
with a direct clarification of how results were generally surmised. A
credit is made here to the Utah Genetics Reference Project, which, though
not thoroughly described, implied meaningful networking of related
scientific projects, potentially providing a subtle suggestion for the
reader to become more aware of and appeciate such outside projects. More
elaboration is provided on the methodology of the study as follows, with a
rationale provided with each step:
"But because ASDs occur in a spectrum of mild to severe symptoms, and the genetic contribution of each of these variants likely varied, they needed to find out which of the 153 aberrations were most strongly linked to autism; some were likely indirectly connected to the disorder, and the scientists wanted to weed out those potential red herrings. So Hensel and his collaborators built a new molecular test, or probe, that would identify the 153 variants from a patient sample of blood, as well as for 185 other gene variants that previous studies had linked to autism. Running this probe on genetic samples collected from 2,175 children with clinically diagnosed autism spectrum disorders and also from 5,801 children with normal development, they could compare how well each of the variants matched up with an ASD."
Rather than merely focusing on what is known or what was discovered, this
passage walks the reader through each of the steps in the process of
discovery, presenting each natural step in the process following each new
result. The probe is presented as the next logical step in the process,
and though it is not discussed in detail, the overall functionality of
this technique is established. The results are presented following the
introduction of this final method:
“Of the 153 initial candidate gene variants, 15 were confirmed as autism-related. The test also picked up another nine autism-related variants that had never been linked to autism spectrum disorders before. Participants with any of the 24 variants had a two-fold greater risk of developing an ASD than those without the genetic changes. Of the previously selected 185 genetic variants connected to autism, 31 were also strongly linked with ASDs, but need additional testing to establish how predictive they are of the disorder.”
No discussion is provided as to exactly how the genes were distinguished
as “related” or “linked” to autism from the results of the technique using
the “probe,” though such a discussion of statistical methods would not be
of much practical relevance to a non-specialist reader. In addition, one
component of the primary study's results – the particular CNVs located through
their associated SNVs – were excluded from discussion in the TIME
article, though this aspect of methods and results would likely have not
contributed a great deal to the article's overall message. The mention of
a need for additional testing creates an impression of how the scientific
process is typically an ongoing process, putting the research in a future
context; additional commentary is made in this vein:
“Autism researchers suspect that as many as 200 to 400 genes may be responsible for ASDs, and that patients will exhibit a wide array of diverse genetic combinations of these DNA contributors. Not every person with autism will have each and every autism-related gene variant, but it’s likely that he will show some variation of the genetic changes identified in the study. “The bottom line is that autism simply is a lot of different diseases genetically, each of which gives you a similar behavioral result. But they have different causes,” he says.”
Though such a statement of how “not every person with autism will have
each sand every autism related gene variant” may seem fairly obvious to a
specialist familiar with the varieties of autism, this point of
clarification seemed quite a courtesy for a lay-reader, as autism is quite
exceptional from many more popularly understood common diseases brought
about by single discrete causes (bacteria, smoking, single gene mutations,
etc.). This provides a way of reaffirming a key message the article wished
to portray. The article concludes with a revisiting of the social and
clinical relevance of the discovery, an effective message for the reader
to consider after finishing the article:
“And that’s why the findings, as encouraging as they are, still can’t be used to diagnose and identify people at risk of developing autism. However, Hensel hopes that large studies like the current one may start to isolate the true genetic predictors of the condition. ‘Because of the size of this study, it really gets us to the cusp of using [genetic markers] clinically,” Hensel says. “We wanted to do a big enough study that we could say, yes, these results really mean something.’ And hopefully, within a few years, they will.”
A study such as this one without direct clinical applications emerging
from it may be a challenge to place in a context perceived as relevant to
society at large. This article does well in concluding with a quote from
study author Chuck Hensel, addressing the predicted importance this study
will have in future studies. The writers of the article also make what can
be assumed a welcome change for the lay-reader in changing what was likely
a more technical description to the bracketed “genetic markers.”
Conclusion
The popular press has an often challenging set of duties in regard to science, acting simultaneously to identify relevant advancements in science, translate these advancements into language relatable to the average person, and deliver the knowledge of how exactly to act on the information provided [3]. The TIME magazine article reported on herein provides an example case study of how the media should approach these duties. It presented a study using abstruse methods in a relevant social context, walked the reader through the process of discovery (thus allowing the reader to acquire the critical thinking "tools" utilized throughout the scientific process), placed the study in past, present and future contexts, discussing which pieces of the puzzle of ASDs remain to be filled and how the study's findings will relate to those findings.
This particular paper also demonstrated a compromise that must be made in reporting scientific findings by providing relatively brief accounts of methods and their general utility used in the study for the sake of keeping the article to a manageable length. Other links were provided within the article itself, however, to satisfy curiosities that the writers suspected that may remain unfulfilled in the reader by this article alone.
References
[1] Autism
and Developmental Disabilities Monitoring Network Surveillance Year 2006
Principal Investigators, Centers for Disease Control and Prevention
(CDC) (2009) Prevalence of autism spectrum disorders - Autism and
Developmental Disabilities Monitoring Network, United States, 2006. MMWR
Surveillance Summary, 58, 1–20.
[2]
Matsunami, N., et al. (2013) Identification of Rare Recurrent
Copy Number Variants in High-Risk Autism Families and Their Prevalence
in a Large ASD Population. PLOSone.
[3]
Kua, E., Reder, M. & Grossel, M. J. (2004). Science in the News: A
Study of Reporting Genomics. Public Understanding of Science, 13(3),
309-322.
[4]
Blue, L. (2013). New Gene variants Linked to Autism. TIME:
Health and Family. Accessed 29 Jan. 2013.
Genomics Page Biology Home Page
Email Questions or Comments to gasmith@davidson.edu.