This web page was produced as an assignment for an undergraduate
course at Davidson College.
Synthetic Yeast Chromosome
Summary and opinion
The successful synthesis of the Mycoplasma mycoides genome
in 2010 was the first time a living cell contained a lab-made genome.
The bacterium, “Synthia,” was touted as the first synthetic life form.
The genome was a technical proof-of-concept, demonstrating that
large-scale DNA synthesis is possible. Though encoded in the Synthia
genome were a few names and quotes (one of which led to the threat of a
breach of copyright lawsuit), there was no genomic novelty that could address basic research questions.
The synthetic Saccharomyces cerevisiae
chromosome III, or synIII, embodies a different scientific approach.
The authors integrated the goals of developing new technical approaches
with establishing an experimental system that can be interrogated for
useful data from many angles. Their thoughtful in silico design for a
streamlined chromosome, containing alterations such as removal of
introns, was successfully made a reality by an army of Johns Hopkins
undergraduates. The chromosome-wide alterations do not appear to affect
fitness in laboratory growth conditions.
synIII is a novel approach in the field of synthetic biology.
Precedents for multiplexed, chromosome scale manipulations are scant,
owing to the complexity of making iterative genome alterations. Through
its integrated recombination sites, synIII could be the first of a new
type of experimental genomics in the spirit of the genetic screen. What
genomes (combinations of genes) can give rise to a phenotype, the most
obvious being viability? The main text of the paper reports an
extremely high rate of recombination at the MAT
locus under induction of their engineered recombinases in diploid
yeast. Recombinase activity was sufficiently high that Cre was toxic to
haploid cells due to excessive gene deletion. An optimized system that
uses a weaker Cre variant could be immensely useful in generating
libraries of mutant, viable yeast lacking specific genes.
Figure 1
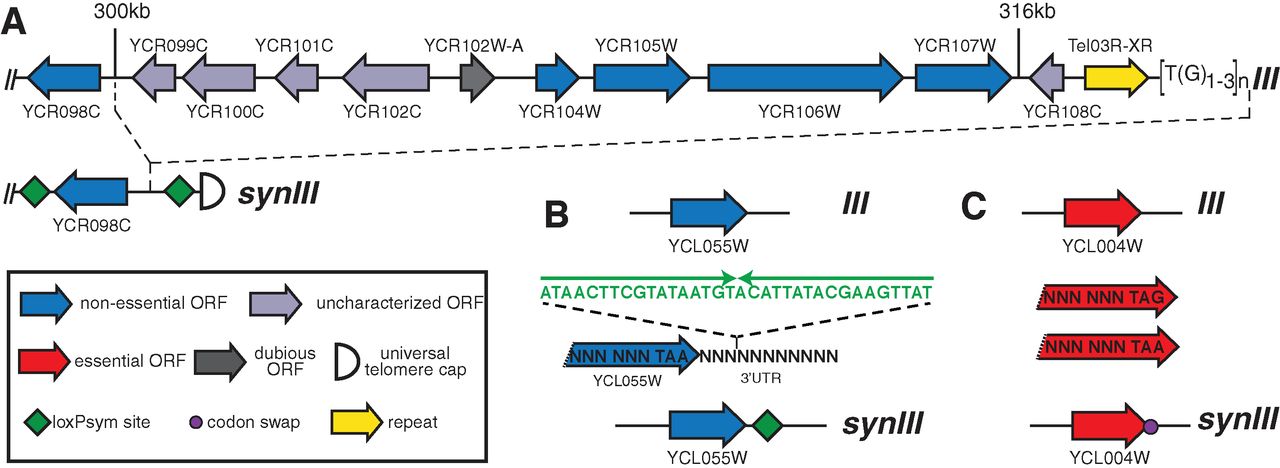
The synIII chromosome is derived from the S. cerevisiae chromosome
III, but with deliberate changes. Figure 1 shows examples of the
changes made across the chromosome. First, all TAG stop codons were
changed to TAA. The elimination of one of the 64 codons from the genome
could be useful in future applications, such as introducing of a fully
orthogonal tRNA carrying a non-standard amino acid. The authors also
integrated loxPsym recombination sites (sym for symmetrical) flanking
genes that are singly non-essential. The loxPsym sites could allow
future researchers to discover large combinations of non-essential
genes. Other genome changes include deletions of introns and tRNA genes
as well as the incorporation of markers (PCRTags) that distinguish
synIII from the endogenous chromosome III.
Figure 2
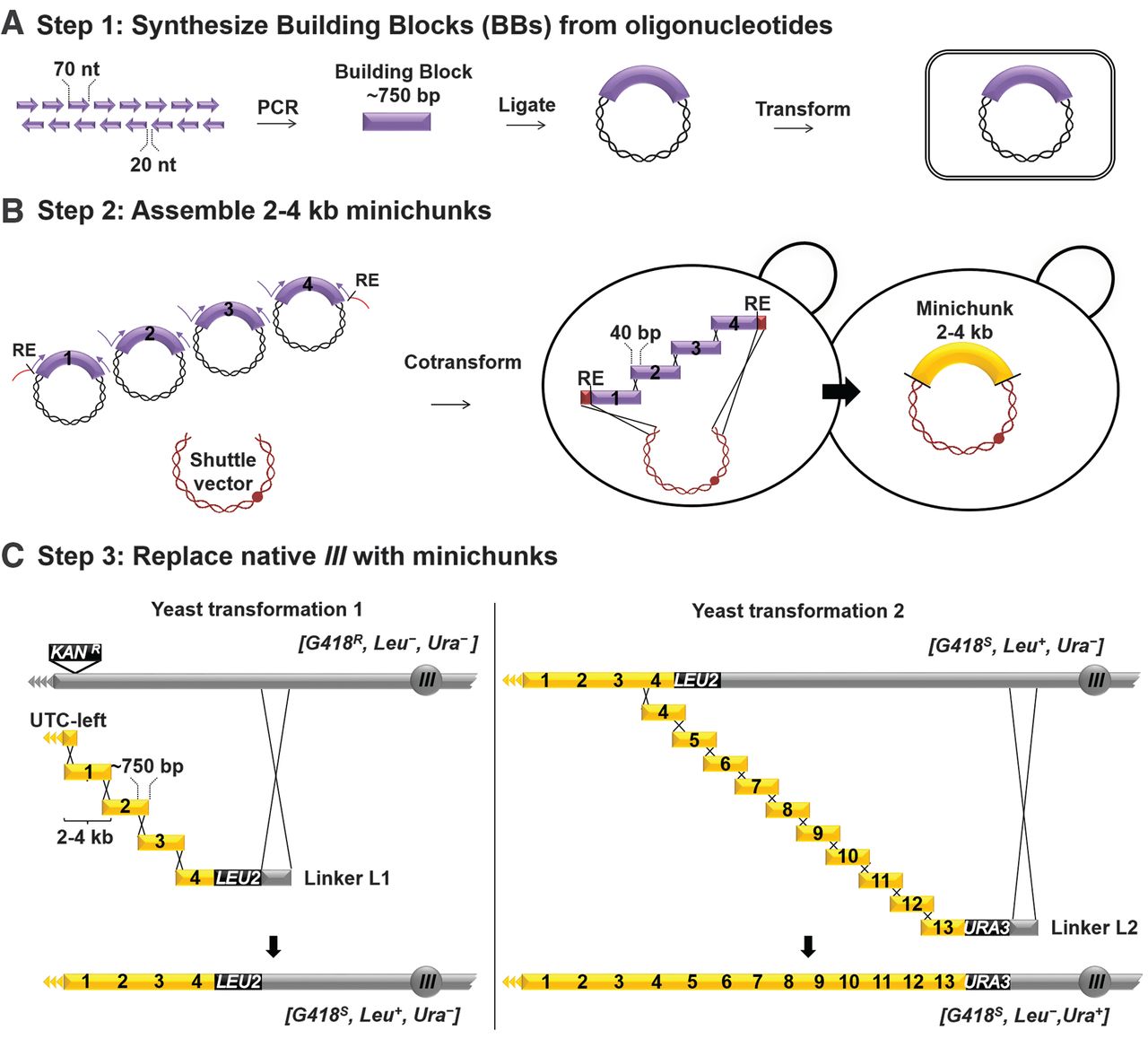
Undergraduates at Johns Hopkins University were responsible for the bulk of synIII assembly. Because in vitro
DNA synthesis produces short ssDNA oligos, the assembly of a complete
chromosome required multiple stages of hierarchical assembly. In the
first step, overlapping oligos were amplified by PCR to produce ~750-bp
dsDNA Building Blocks. Building Blocks were then assembled into
Minichunks by homologous recombination in yeast, facilitated by short
overlaps between adjacent Building Blocks built into their design.
To generate the fully assembled synIII, they used homologous
recombination to swap segments of the endogenous yeast chromosome III
with their synthetic chromosome. By alternating between the selectable
markers LEU2 and URA3,
they incorporated synIII segments in a stepwise fashion along the
length of chromosome III. The successful integration of each set of
Minichunks is associated with flipping between Leu−/Ura+ and Leu+/Ura− phenotypes.
Figure 3
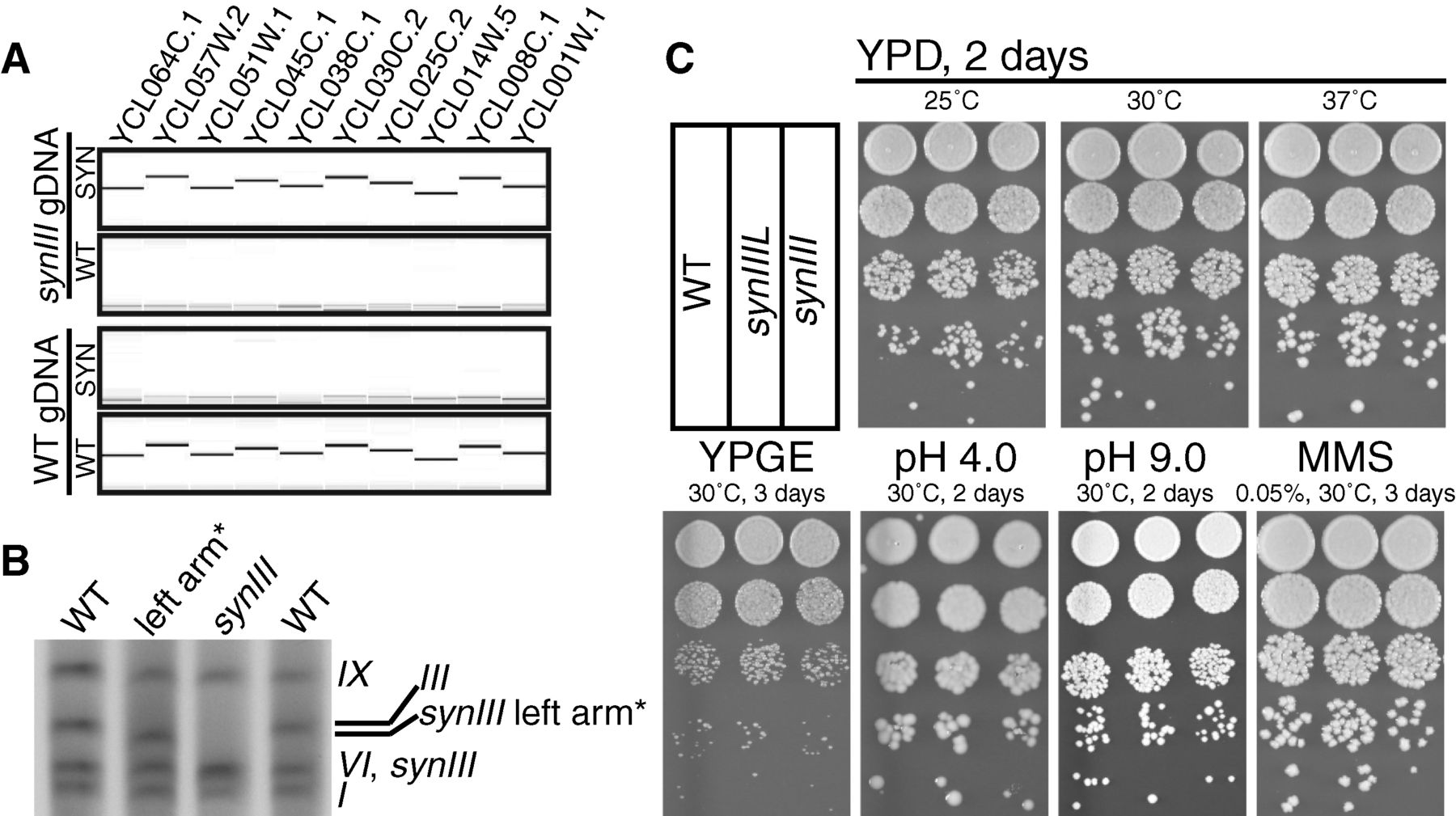
To ensure that the endogenous chromosome III was fully
converted to synIII by their homologous recombination scheme, the
authors used PCRTags (markers) unique to either the wild type
chromosome or synIII. Using gDNA extracted from wild type and synIII
strains, they found that the PCRTags only generated a PCR product when
matched with their cognate strain. The result indicates that the wild
type chromosome was successfully converted to synIII in the engineered
strain. By running the gDNA on a gel, they confirmed the smaller length
of synIII relative to III by increased migration distance through the
gel matrix. Genome sequencing (not shown in the figure) indicated
almost complete replacement of the wild type chromosome with synIII
sequence, except for 10 sites, mostly single bases. Additionally, they
tested growth phenotypes for synIII and the assembly intermediate
synIIIL under different culturing conditions. They found no differences
in colony morphology or titer between the two engineered strains and
wild type under the various growth conditions.
Figure 4
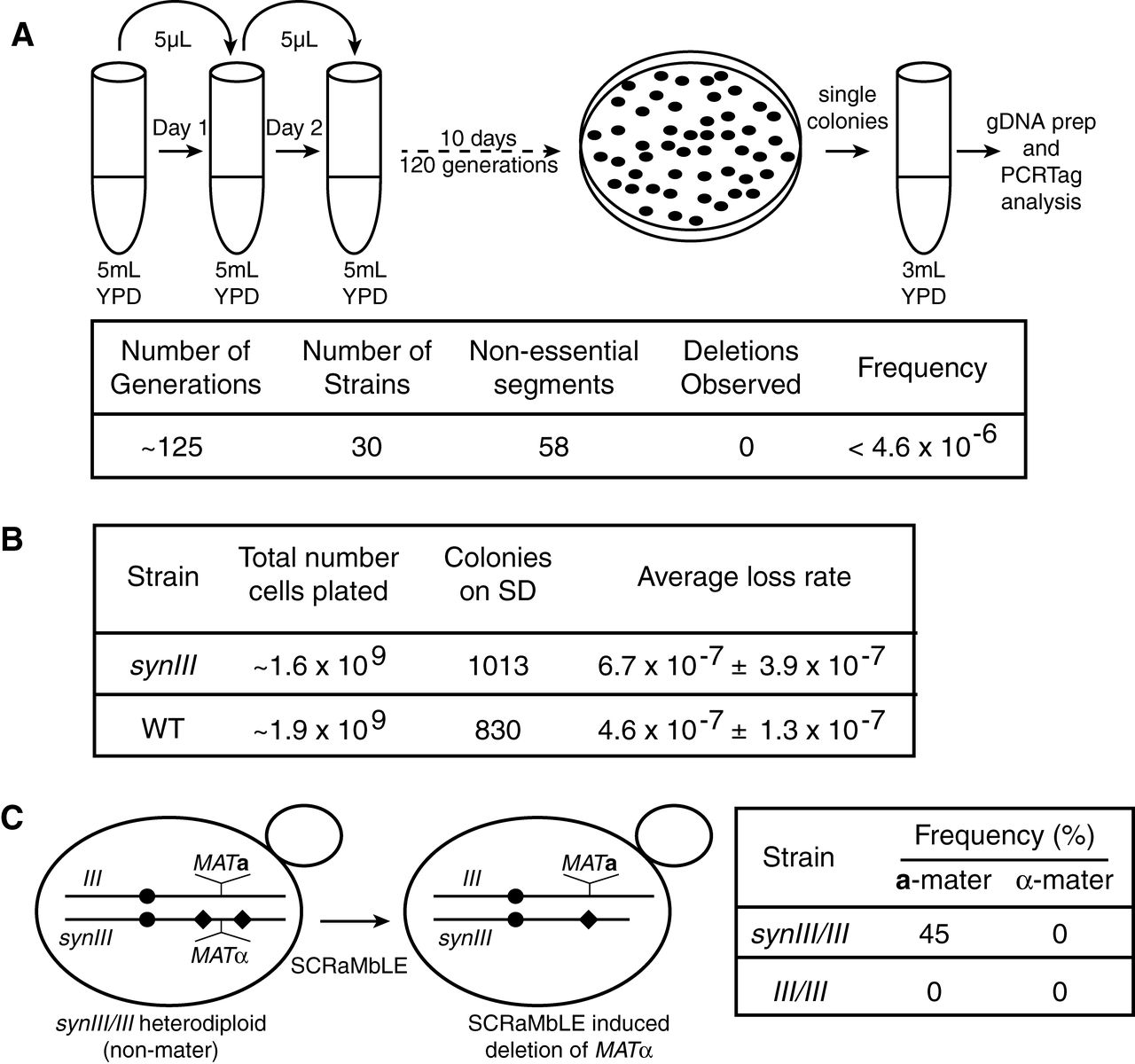
A whole genome transcriptome analysis found differential
expression of only two genes present in both synIII and wild type,
indicating that the chromosomes are functionally similar. The authors
also monitored the stability of synIII over 125 generations using 30
replicate strains by tracking their PCRTags, finding no deletions. They
also measured the frequency of conversion to the MATa mating type from MATα, an indication of chromosome III loss due to the location of MAT
on chromosome III. Consistent with their PCRTag result, there was no
significant difference in mating type conversion rates between synIII
and wild type. In the final experiment of the figure, they expressed
Cre recombinase to introduce random deletions using the loxPsym sites
included in synIII, a process they called SCRaMbLE. In heterozygotes
carrying the MATα allele on synIII and the MATa
allele on wild type III, almost half of the yeast became mating type a.
The wild type diploid heterozygotes are normally non-mating. Expression
of Cre in MAT heterozygotes
carrying only wild type chromosomes failed to produce mating diploids,
as expected. PCRTag mapping confirmed that mating diploids carrying
Cre-edited synIII lost the MATα locus, allowing the yeast to adopt the a mating phenotype.
Reference:
Annaluru N and Muller H et al. 2014. Total synthesis of a functional designer eukaryotic chromosome. Science 344:55-58.
Eric Sawyer's Home Page
Genomics Page
Biology Home Page
Email Questions or Comments.
© Copyright 2014 Department of Biology, Davidson College,
Davidson, NC 28035