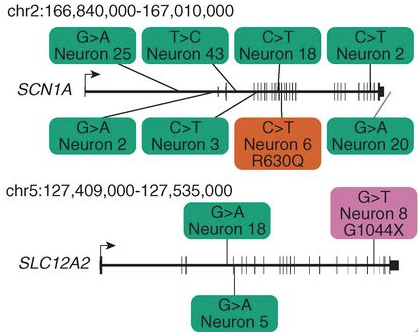
Figure 1. Two genes implicated in human diseases from brain B with SNVs. Green boxes represent SNVs that occured in introns, the orange box represents an SNV that ocurred in an intron resulting in a missense mutation, and the purple box represents an SNV that caused a nonsense mutation.
Source: Science, AAAS*
The researchers compared the SNVs they found across all 36 neurons and found that depending on the similarities, they could determine which SNVs came first, thus determining a preliminary lineage tree. For example, if a certain SNV was found in many of the neurons of a the brain, that SNV was probably older, whereas, if only a few cells have a certain SNV, that SNV is probably newer.
Lodato et al. also sequenced tissues from other parts of the body such as the heart and found that these cells also shared some of the same SNVs as the neurons. This led them to believe that these SNVs occurred before the tissues separated at gastrulation, and likely came from the same founder cell (Lodato et al., 2015).
Discovery Science:
This study sought to examine and analyze the presence of somatic SNVs in the human brain. Although they did not set out with a specific hypothesis, their results regarding the developmental and transcriptional history of cells open up various hypotheses to test regarding exploration of somatic mutations and their relationship to brain disease.
Genomic Technologies Used:
Lodato et
al., within their large single-cell whole-genome sequencing
project used several methods to not only get data from the neurons
they tested, but also to verify their results. First,
fluorescence-activated nuclear sorting (FANS) was used as a way to
study the nuclei of the neurons while preserving post-translational
modifications. The DNA was amplified using multiple-displacement
amplification (MDA), a procedure similar to PCR, with the advantage
of lower error rates, but the disadvantage of artifactual deletions
(Lodato
et al., 2015).
The DNA was then subjected to high-throughput sequencing. Using three different mutation-calling algorithms, the investigators identified single-cell SNV candidates, and then confirmed these results with Sanger sequencing (Lodato et al., 2015).
Take Home Message:
This study has implications
regarding disease, developmental history, and the gene expression in
neurons themselves. One important finding was that mutations in
neurons do not arise due to replication as in cancer. Rather, the
locations of the elevated mutation rates (at transcribed loci and
DNase I hypersensitivity sites) suggested that the vulnerability is
caused by RNA transcription and histone dissociation (Linnarson,
2015). This implies that mutations arise, not when DNA is
replicated, but rather when it is unwound and transcribed. This
could mean that the more a gene is expressed, the greater the
chances are of it being mutated (Howard
Hughes Medical Institute).
As demonstrated in Figure 1 above, this study also shows that the idea of carrying out single-cell whole-genome sequencing could help to identify somatic mutations involved in brain disease (Linnarson, 2015). This small-scale study demonstrated how from sequencing just a few cells, a preliminary cell lineage tree of an individual could be inferred. Whole-genome sequencing is a fairly expensive process and applying this procedure to just single-cells is just expensive as it would be to sequence an individualís entire genome. Thus, although the resources and funding for such an endeavor may be out-of reach at this time, this study shows that theoretically, it would be possible to deduce the lineage of every cell in an individuals body using a similar method of identifying somatic SNVs and mutations.
Evaluation of Project:
Overall, this project seems to have provided several new insights into the implications of mosaicism in the brain that also have ties to various other cells in a personís body. The research conducted by Lodato et al. opens doors to new possibilities of cell lineage exploration. It shows how the work that has been done so far gives rise to new actionable hypotheses. At the same time, the high costs of replicating and repeating the procedures used remain a hindrance to taking the study to the next level. Until researchers have sufficient resources and funds, it may be difficult to apply the findings of this study to discovering the entire developmental and transcriptional history of an individual. The investigators also suggest that it may be worth it to see if the mutation rate in neurons increases with age, as they recognize that their sample size of three individuals does not provide sufficient data to make any conclusions in that respect. The project was conducted in a thorough manner, taking care not to take the results at face value and instead, running other tests to verify the results. For example, while searching for SNVs, the investigators used three different methods and then confirmed these findings at a rate of 92% using Sanger sequencing. Thus, it not only presents data in support of its claims, but also makes it difficult to refute or disagree with their findings due to the thoroughness of their research.
*Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.
Citations/Sources:
Howard Hughes Medical Institute. Single neuron may carry over 1,000 mutations. ScienceDaily [Internet]. [Last updated 1 October 2015, cited 30 Jan 2016]. Available from: www.sciencedaily.com/releases/2015/10/151001153931.htm
Linnarsson, S. 2015. A tree of the human brain. Science [Internet]. [Last updated 2015 Oct 1, cited 2016 Jan 30] 350(6256):(37). Available from: http://science.sciencemag.org/content/350/6256/37
Lodato, M. A., Woodworth, M. B., et al. 2015. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science[Internet]. [Last updated 2015 Oct 1, cited 2016 Jan 29] 350(6256):(94-98). Available from: http://science.sciencemag.org/content/350/6256/94.full
Email Questions or Comments: saayyar@davidson.edu.
© Copyright 2016 Department of Biology, Davidson College, Davidson, NC 28035