This web page
was produced as an assignment for an undergraduate course at Davidson
College.
Review
of:
The phenotypic legacy of admixture between modern humans and
Neandertals
Summary
The study by
Simonti et al. (2016) sought to determine whether the introgression of H.
neanderthalensis into H.sapiens genome could have an
impact on the variation of clinical phenotypes in a modern
European-descent population. The research consisted on pairing phenotype
data publicly available on electronic health records (EHRs) from the
Electronic Medical Records and Genomics (eMERGE) Network, which combines
patientsí genetic data from nine cities in the U.S.
28, 416 patients of
European descent were pooled from the set, and a set of over 135,000
SNPs most likely derived from Neandertal alleles was filtered. Previous
studies had hypothesized a correlation between Neandertal alleles and
variation in lipid metabolism,immunity, depression, digestion, hair, and
skin. Thus, the cohort used genome-wide complex trait analysis (GCTA) of
1495 Neandertal SNPs, and found that the risk of depression, myocardial
infarction, and corns and callosities were significantly explained by
the SNPs (and in their replication as well).
The study then
focused on finding if there was any correlation between single loci and
modern human phenotypes. They performed a phenome-wide association study
(PheWAS) of the same 1495 SNPs, and demonstrated four highly significant
phenotypic associations (and in their replication as well).
Finally,
they also showed that Neandertal SNPs were highly associated with
neurological and psychiatric phenotypes, and with fewer digestive
phenotypes. The cohort alluded that the Neandertal SNPs were probably
advantageous to early modern humans, and these are mismatches due to our
current modern environment.
Opinion
In general, the cohortís methods
and structure efficiently established the rationale for their
investigation, as well as demonstrated tangible and verifiable results
explaining the phenotype association between Neadertal and modern humans.
The second phase based on replication of data sets was a sound approach to
reduce false discovery rates, and further make their conclusive remarks
more significant. Also, I thought that the use of EHR data was interesting
aspect of the paper as it opens possibilities for paired meta-analysis
with other big data sets available, and could further our understanding on
clinical pathologies. However, I had difficulty in grasping that the EHRs
were actually integrated through the eMERGE network. At first, it seemed
that both were used simultaneously to achieve the Neandertal SNPs data
set. Moreover, even though the use of different methods was justified and
successful in establish the cohortís conclusion, the extent to which they
were explained was lacking. Additionally, the statistical and
computational analyses could have had a more detailed background
explanation (without prompting the reader to look at supplementary
materials or elsewhere). Lastly, Figure 1 was particularly difficult to
interpret and follow the full array of the methodologies used, since their
rationale for them is explained in subsequent parts of the text.
Figure Summaries
Figure 1
Overall, the figure demonstrates the array of methods
used in this investigation that lead to their various conclusions. Panel A
shows the pairing of patients' genotype with EHR in the eMERGE Network in
order to look for clinical phenotypes and Neandertal SNPs. Panel B and C
shows the genotypic and phenotypic similarity between the patients in all
Neandertal SNPs, and the use of GCTA in Neandertal SNPs in order to find
the disease-risk associated with them, respectively Panel D shows the PheWAS
study to demonstrate the four highly significant SNPs. Panel E focuses in
one of the significant SNPs and its expression level.
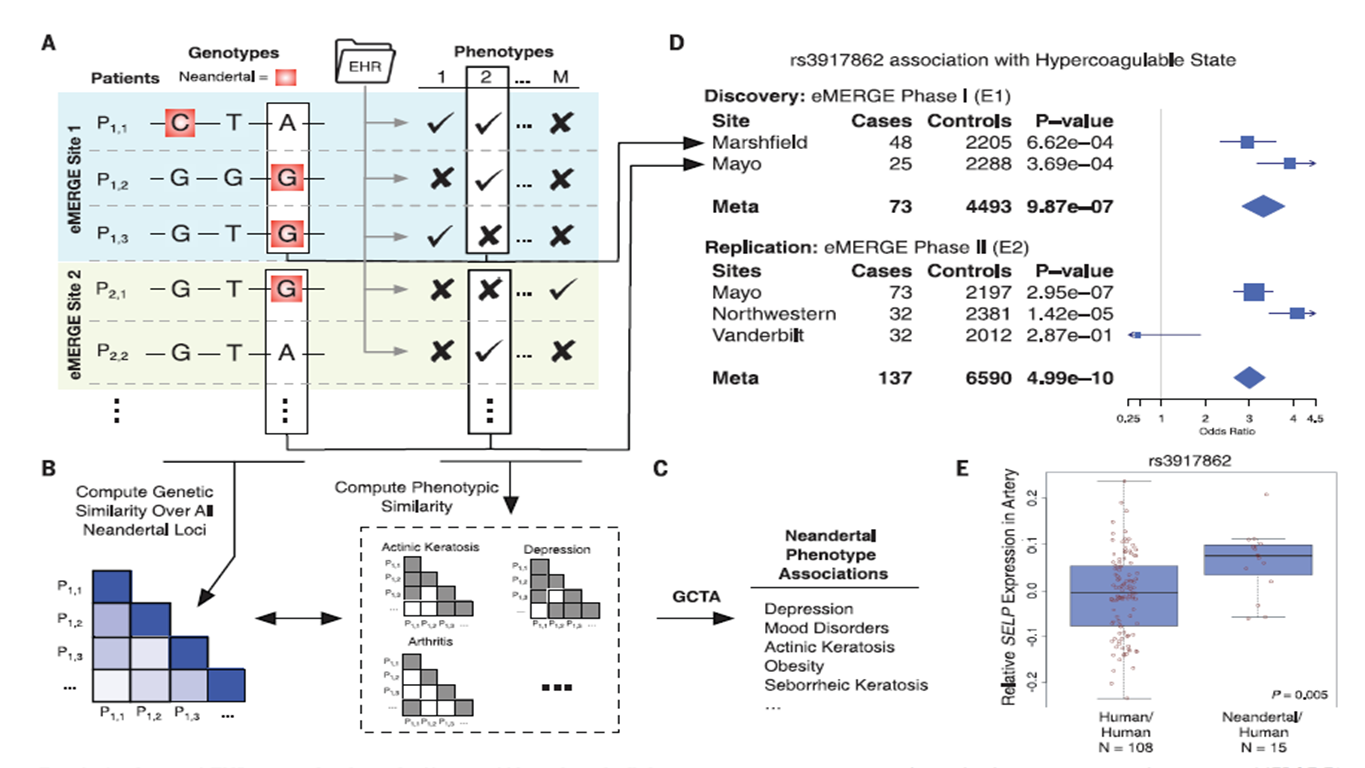
Table 1
This table shows the results of the GCTA, and
demonstrates eight traits that can be associated with Neandertal SNPs in
both the discovery and replication sets. Three traits remained significant
after a GRM analysis, which accounted for non-Neandertal variation:
depression, myocardial infarction, and corns and callosities. The figure
demonstrates the verifiability of their methods, and that their remarks
about phenotype associations are tangible.
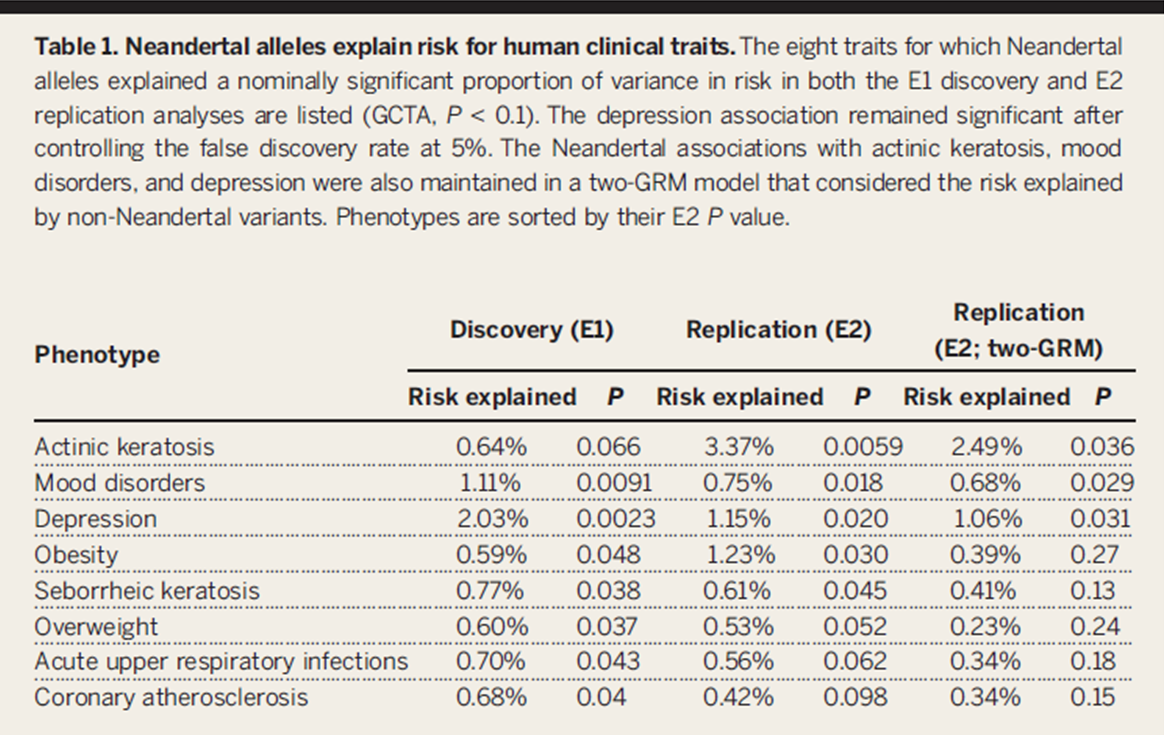
Table 2
This table highlights the four individual significant
Neandertal SNP-phenotypic associations with the use of PheWAS method in
both the discovery and replication set. The figure highlights the importance
of higher scrutiny and replication in order to attain equally verifiable
results.
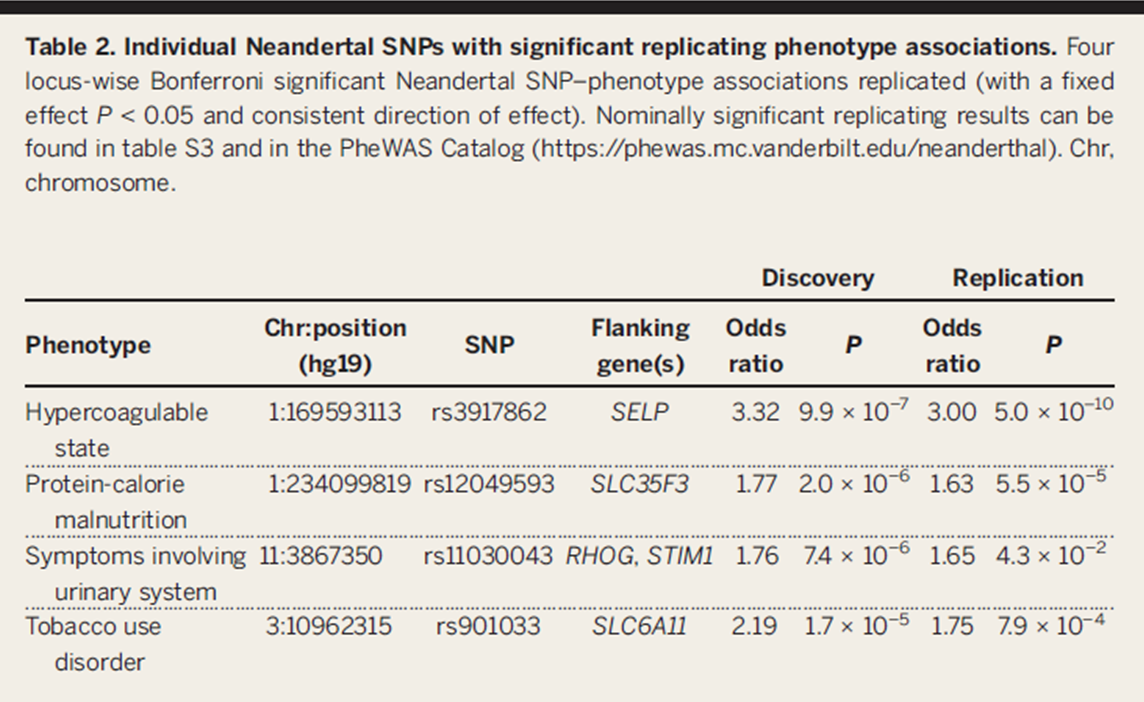
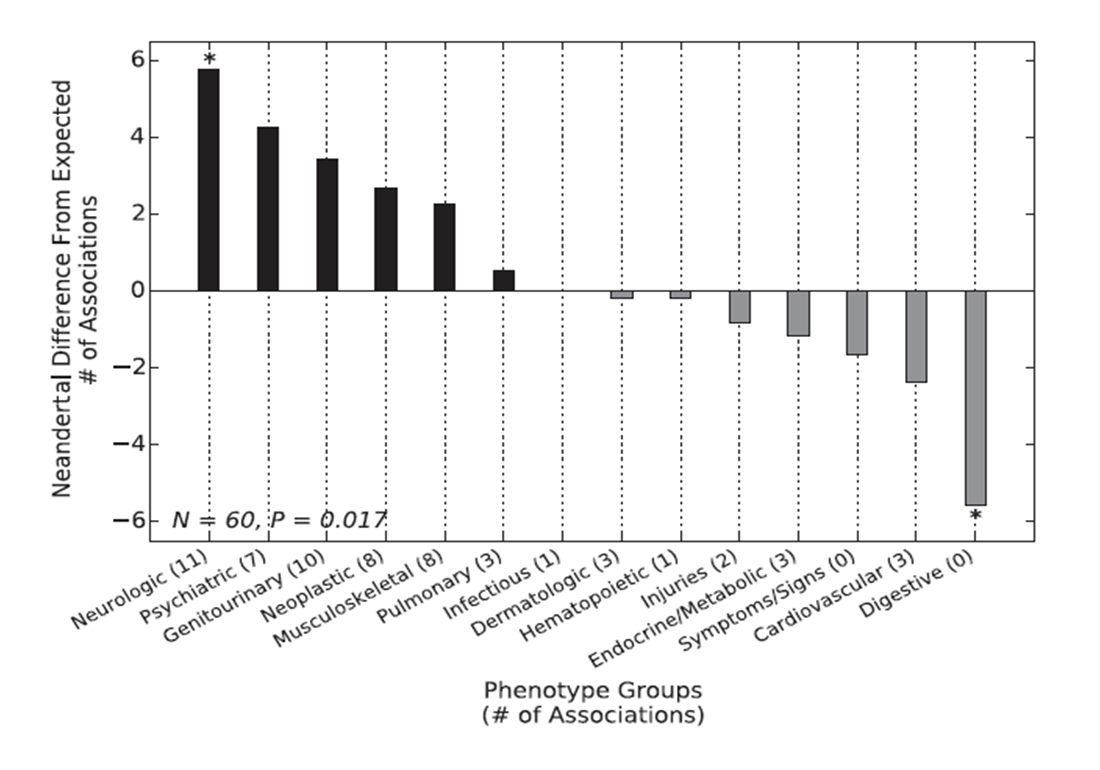
Figure 2
This figure shows the Neandertal SNP enrichment in
association with specific phenotypes using the PheWAS method. By comparing
phenotype-associations between Neandertal and non-Neandertal, they showed
that Neandertal SNPs are more highly associated with neurological and
psychiatric phenotypes than digestive ones.
Reference
Simonti, C, Vernot, B, Bastarache, L, Bottinger, E,
Carrell, D, Chisholm, R, Crosslin, D, Hebbring, S, Jarvik, G, Kullo, I,
Rongling, L, Pathak, J, Ritchie, M, Roden, D, Verma, S, Tromp, G, Prato,
J, Bush, W, Akey, J, & Denny, J. 2016. The phenotypic legacy of
admixture between modern humans and Neandertals. Science 351:737-741. http://science.sciencmag.org/content/351/6274/737
Genomic
Home Page
Genomics
Page
© Copyright 2016 Department of Biology,
Davidson College, Davidson, NC 28035
Send comments, questions, and suggestions to: brmourao@davidson.edu




