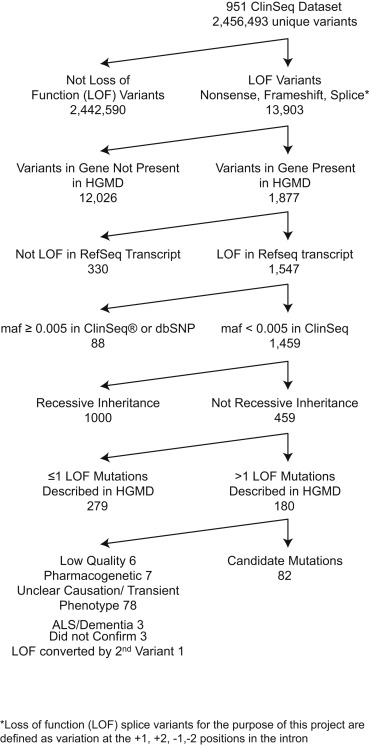
Figure 1. Filtering chart used to detect pLOFs meriting followup via iterative phenotyping. Johnston et al., 2015. Used with permission.
This webpage was created as part of an undergraduate assignment at Davidson College
NIH researchers pilot predictive medicine by studying healthy people's DNA
Q1) Summary:
A team of National Institutes of Health researchers have "turned traditional genomics on its head" (NHGRI, 2015). In a study sponsored by the National Human Genome Research Institute, the National Institute on Deafness and Other Communication Disorders, the National Eye Institute, and the National Institute of Diabetes and Digestive and Kidney Diseases, the team in question sequenced the genomes of 951 healthy individuals, searching for mutations most likely to lead to a genetic disease. About 10% of the volunteers in the study had mutations known as pLOFs - putative Loss-of-Function mutations. A pLOF is predicted to cause a phenotypic change, and the most severe mutations are should be the best at predicting disease (Johnston et al., 2015; Biesecker, 2015). The researchers then applied a method they called "iterative phenotyping": selective clinical follow-ups with the volunteers in whom pLOFs had been identified (NHGRI, 2015). 34 of these individuals had personal or family medical history that was linked to the specific condition predicted by their pLOF (Johnston et al., 2015).

Q2) Genomic Methods and Technologies Used:
The genomes of participants in the study had been previously sequenced using Next-Generation Sequencing as part of ClinSeq, a large-scale NIH study of the use of individualized genome sequencing in clinical settings. The researchers collected and analyzed the exome data of 951 volunteers in the ClinSeq cohort (Johnston et al., 2015). They used global literature to identify which mutations were known to be associated with diseases (Biesecker, 2015).
Q3)
Purpose of the Study:
This was a proof-of-concept study which sought to determine whether it is possible to use genomic analysis find patients who are at risk for illness and begin care before symptoms present themselves. Further studies will involve identifying less pathogenic (less severe) mutations and investigating their influence on health and disease (Biesecker, 2015).
Q4) Take-home Messages:
The
researchers were very positive about the accuracy of their
predictions. In an individual with no known medical history, the
accuracy of predictions of disease was 1 in 2 (50%) when using genome
sequence data, as opposed to 1 in 50,000 using traditional methods.
They also had the opportunity to study individuals with a pLOF
but mild or no symptoms of the associated disease. This
information will help clinicians and researchers better understand
other factors that influence whether or not disease symptoms appear.
34 of 951 apparently healthy individuals involved in the study were
found to have a genetic condition. This statistic suggests that
genetic disorders are more common than previously thought. The
commonly accepted estimate has been less than 0.02% of the US
population may have such a condition, but in fact this study suggests
that 3% or more of Americans may suffer from some genetic condition.
The vast underestimate is understandable - most genetic disorders are
discovered because of family health history or ill people looking for
a diagnosis. Since individuals exhibiting mild or no symptoms, such as
20 of the 79 participants who never thought their physical change
could be linked to a genetic cause, often do not seek diagnosis or
treatment, their experiences cannot be studied. As such, genetic
diseases have a reputation for having a lower prevalence and greater
severity than may be the case. It is important to be aware of the
possibility of silent disorders, because a mutation that causes only
mild symptoms in a parent may have dangerous consequences in the
offspring (NHGRI, 2015).
However, the researchers warn that the results of this study do not
mean that every apparently healthy person should rush to have his or
her genome sequenced. They wanted to show that genome sequencing can
be useful in predicting illness even in those without severe symptoms,
or before symptoms present themselves. Their goal is to "add DNA
sequencing to the physician's toolbox" as a diagnostic tool and also a
preventative medicine tool when family history indicates it is
appropriate (qtd. in NHGRI,
2015).
Q5) Analysis:
The results of this study clearly have very real implications for medicine. It is amazing that genomic sequencing can be such a powerful tool, to the point of indicating serious underlying diseases in people with only mild symptoms. The example of individuals who thought they had basic high cholesterol and actually suffered from familial hypercholesterolemia is a strong case in point. This is a very severe condition and so it would be useful if a physician who is advising a patient on how to deal with high cholesterol could screen the patient for this disease. As noted in the article, this could have powerful results not only for the patient but for the family members, who may also have the disease (NHGRI, 2015).
I was particularly intrigued by the new estimate of the prevalence of genetic conditions. It makes sense that those who experience the most severe symptoms will be the ones to seek treatment, and thus that the diseases will appear to be rare and serious. This could have serious negative consequences! I imagine if I had mild symptoms of a genetic disease but didn't bother to seek treatment because available information says that my symptoms would be much more severe if I had the condition, and anyway it is very rare. Then I could pass this disease on to my children who could have much more serious symptoms and I would not have anticipated having to care for them. Three percent of the population is quite high. If Davidson were the population, sixty students would suffer from such a disease. Sixty may not sound like that much, but if you consider the whole of the United States, the entire state of North Carolina could have a genetic condition!
I feel that the project was designed well. The practice of iterative phenotyping was wise. It clearly demonstrates that based on genomic information alone, disease can be identified. It also saved them time because they knew which disease(s) to screen for in each individual. If they had started by looking at phenotypic evidence, they would have been subject to the same ascertainment bias which skews estimates of the prevalence and severity of genetic conditions. By starting with genotype, they show that DNA sequencing can be a critical and necessary diagnostic tool in clinical settings. I would be interested to learn what the ultimate predictive power of this practice is, once the researchers attempt to scale it up and test less severe mutations that may be harder to find or less indicative of disease.
Citations:
Beisecker LG. 2015. NHGRI Pilot Project Uses Genomic Sequence Data to Do Predictive Medicine [video on Internet]. Bethesda(MD): National Human Genome Research Institute; [cited 2016 Jan 2014]. Available from: https://youtu.be/GRX85vaVaUI
Johnston
JJ, Lewis KL, Ng D, Singh LN, Wynter J, Brewer C, Brooks BP, Brownell
I, Candotti F, Gonsalves SG, Hart SP, Kong HH, Rother KI, Sokolic R,
Solomon BD, Zein WM, Cooper DN, Stenson PD, Mullikin JC, Biesecker LG.
2015. Individualized Iterative Phenotyping for Genome-wide Analysis of
Loss-of-Function Mutations. The Am. J. of Human Genetics [Internet].
[Cited 24 Jan 2016] 96(6). Available from: http://www.cell.com/ajhg/abstract/S0002-9297%2815%2900150-0
NIH
researchers pilot predictive medicine by studying healthy people's DNA
[Internet]. 2015. Bethesda(MD): National Human Genome Research
Institute (NHGRI); (cited 2016 Jan 24). Available from: https://www.genome.gov/27561786.
Genomics
Page
Biology Home
Page
Email Questions or Comments: moprudencio@davidson.edu.
© Copyright 2016 Department of Biology, Davidson College, Davidson, NC 28035