This web page was produced
as an assignment for an undergraduate course at Davidson College.
Anthony Ciancone's Genomics
First Assignment Page
'Undead' genes come alive days after
life ends
Click
here to view the article. Alternatively, click
here to view the original paper's source.
1) What was the research
project?
The article I read for my first assignment was entitled "'Undead' genes
come alive days after life ends" and was written by Mitch Leslie.
According to the article, Dr. Peter Noble of the University of Washington,
Seattle, and his colleagues were testing new methods for calibrating gene
activity measurements on recently deceased species, specifically on mice
and zebrafish. They were also observing any notable changes in gene
activity in these deceased animals up to four days postmortem.
2) Were they testing a
hypothesis or doing discovery science?
Noble and his colleagues were primarily intrigued to see how well their
methods worked on these deceased animals, as Noble stated: "Itís an
experiment of curiosity to see what happens when you die." However, Noble
and his team were also further testing how well their novel analytic
technique worked. They did not have a specific hypothesis to test but
rather had an interest in seeing how they could apply their new
methodology.
3) What
genomic technology was used in the project?
Nobel and his colleagues claimed to be using new methods for calibrating
gene activity measurements, but did not delve into much further detail.
Previous experiments utilizing the blood of human cadavers had identified
a handful of genes but Noble and colleagues analyzed over 1000 using their
novel methodology. They tracked any genetic changes over a two day period
for the mice and a four day period for the zebrafish. The discoveries
included many genes that not only increased activity up to 24
hours after death but even some that remained active for up to four days,
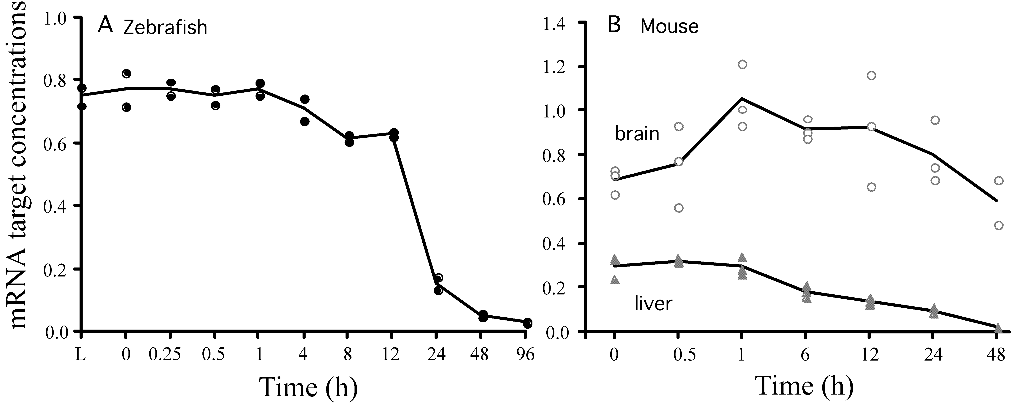
as shown in Figure 1.
Figure 1. mRNA activity vs. time
passed in hours. According to the study, this was looking at genes which
were specifically abundant in mRNA production
postmortem. The y-axis is an arbitrary unit comparison to standard or
expected mRNA production from these genes in living controls. The x-axis
is simply how
much time passed from when they started recording the data, not from
when the organism died; however, these two times were relatively close
when compared to
the overall time frame.
Although many of the genes identified were
related to inflammatory and immune responses, the surprising discovery was
that many 'developmental genes' turned on after death. These
'developmental genes' were thought to be used to help sculpt the embryo
and not used again after birth. Not only that but many genes known to
cause cancer saw a similar increase in activity.
While the researchers grappled over theories as
to the purpose of such gene expression, some potential uses
proved immediately clearer. According to the researchers, the cancerous
genes' increased potency could explain why organs received from deceased
donors predispose the receiver to getting cancer. In a
subsequent paper, Noble claims that observing these genes could
provide forensics teams with the ability to pin point time of death more
accurately. Further uses may not be as apparent but further time and
research may elucidate some of these facets.
4) What was the take home
message?
The take home message from this article is that we can learn a lot about
genes from studying not only living organisms but also deceased ones.
However, many questions remain as to what this research really means. Why
are embryonic genes activated after death? Could these findings lead to
better donor matches? Is this study translatable to humans, or is it
restricted by its dependence on mice and zebrafish? It appears that more
questions remain than this study provides answers for.
5) What is your evaluation of
the project?
My evaluation of this project is that this is a very intriguing finding
and other research should be done to not only confirm the findings but
also assess its utilities, especially when applied to humans. I would be
more interested in learning about the specific genomic techniques utilized
by Nobel and colleagues. How exactly do they differ when compared to
contemporary methods? Why would these methods be best suited for this
project? Another question I had was why Noble and his team did not simply
study cadavers to immediately ascertain applications for humans. They draw
many comparisons but do not address this shortcoming. Overall, however,
this study was very well-done and did a terrific job at assessing an array
of genes and their activities over a specific time frame.
References:
Leslie, Mitch.
"'Undead' Genes Come Alive Days after Life Ends." Science | AAAS.
Science, 06 July 2016. Web. 02 Feb. 2017.
Pozhitkov,
Alexander E., Rafik Neme, Tomislav Domazet-Loso, Brian Leroux, Shivani
Soni, Diethard Tautz, and Peter Anthony Noble. "Thanatotranscriptome:
Genes Actively Expressed after Organismal Death." Thanatotranscriptome:
Genes Actively Expressed after Organismal Death | BioRxiv. BioRxiv
Beta, 11 June 2016. Web. 02 Feb. 2017.
This is my first
assignment homepage. Click here to return to
Anthony's Genomics homepage.
Genomics
Page
Biology Home Page
Email Questions or Comments: anciancone@davidson.edu
© Copyright 2017 Department of Biology,
Davidson College, Davidson, NC 28035