This web page was produced
as an assignment for an undergraduate course at Davidson College.
Review: A global reference for human genetic variation
The 1000 Genomes Project
Article
Link

Figure
borrowed from: Genetic
Literacy Project
Background
The
1000 genomes project initial phase begun in 1990 as part
of an international collaboration of researchers from Germany, China, the
United
Kingdom, and the United States. By sequencing the genome of 1000 people,
this
project produced an “extensive catalog of human genetic variation.” The
Human Genome Project had great success with data generation, storage, and
analysis. With this
data we have already advanced
understanding of disease biology and the processes that shape genetic
diversity.
Part of the 1000 genomes project included the production of a series of tutorial
videos to provide guidance to researchers who want to
access the project's data. Although the official 1000 genomes project is
finished, publicly
accessible data in combination with current research could
continue
to provide solutions to genetic human diseases.
Evaluation of project
I really enjoyed the content of this paper. For the most part, the writing
was clear and did not have extraneous jargon. The authors did a fantastic
job of covering an enormous amount of information in just 6 pages. However,
because this paper references 13 supplemental and 124 of extended
information pages, it's often difficult to fully interpret and establish the
same conclusions that the authors make without spending long periods of time
searching for the supplemental figures. One figure that would have been
beneficial to include (the first figure posted below on this page) shows the
descriptions as well as letter and color code for all 26 sampled
populations. In each figure, the code and color of each population is held
constant, and its a critical aspect to analyzing the presented data. If this
supplemental figure was included, it would make it easier for readers to
follow patterns.
One of the main factors I appreciate about this article, is the clear
acknowledgment of previous genetic bias in genetic studies and the conscious
effort this group made to sample people from across the world. However,
given that the human reference genome is primarily composed of people with
European ancestry, other groups will always appear to have a greater degree
of variance. For example, if the human reference genome was primarily
composed of people with African ancestry, Figure 1B would look the opposite
to what it does now. African groups would have the least variation, and
those of European groups would have the most variation.
Overall, I predict that the information presented in this article will be
the basis of extensive work for years to come. Establishing a way to
determine the phenotype of all variants in all humans which can lead to a
more comprehensive way to look at personalized medicine. Since this group
has collected and processed information from 26 populations, this approach
will not only look at a person's ancestry and determine a European, Asian,
or African drug, but instead will be able to determine the exact medication
a single person and their exact genetic markers need.
·
Explanation of Figures
Supplemental Figure
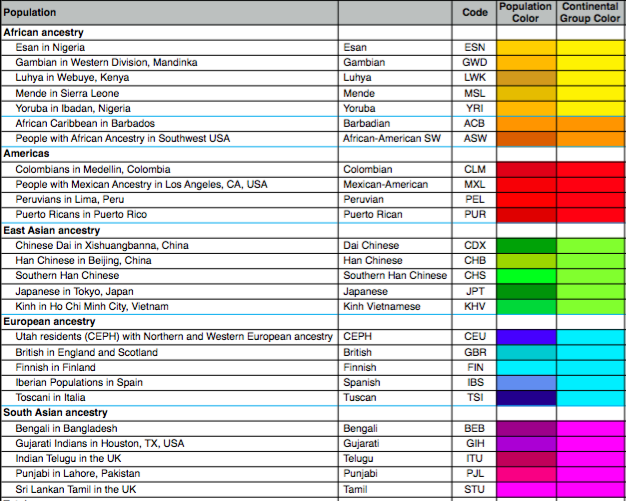
Supplemental Figure 1. Description, letter code, and color code for
26 worldwide populations.
Article Figures
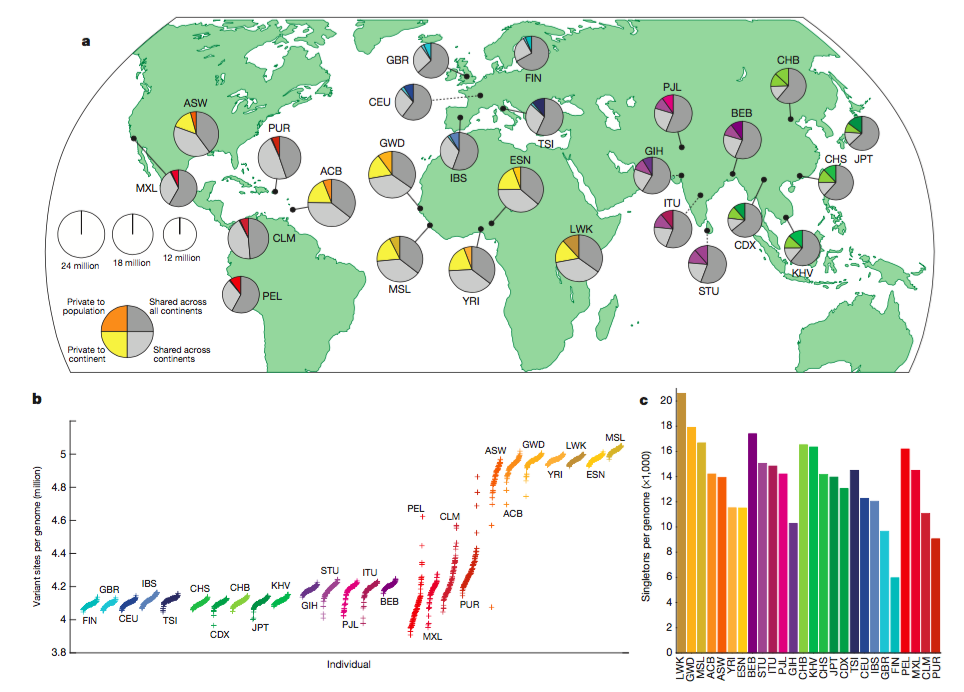
Figure 1
A. Twenty-six
populations throughout the world were
sampled. Each person’s genotype, haplotype and genetic variation was
estimated
by whole-genome sequencing, targeted exome sequencing, and high-density
SNP
microarrays. Each pie chart represents one population, and each color
within
each pie chart represents the variation of that population. Grey indicates
continental variation: Variation present in all continents (Dark grey) or
variation only across continental
areas (light grey). The population specific color represents variation
private
to population (dark population specific color) or variation private to
continental area (light population specific color). Area
of chart is indicative number of
polymorphisms within the population.
For all populations, the
greatest amount of
variation is shared between continents.
B. The number
of variant sites (SNPs, indels, and
structural variants) in an individuals genome as compared to the human
reference genome. Since the human reference genome is primarily
composed of European genes, individuals
with
European ancestry (FIN, GBR, CEU, IBS, TSI) have the fewest variant
sites and
individuals of African ancestry have the most variant sites per genome.
C.
Singletons (variants observed in only one
population) for all populations constitute a very small portion of all
variant
sites per genome.
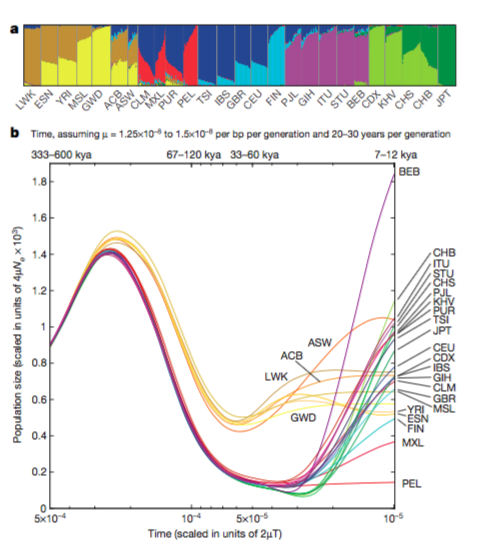
Figure 2.
A. The
proportion
of an individuals genome from putative ancestral populations computed
using a maximum likelihood approach. Each column represents a human
sequence. Ordering
of columns is first done by similarity within a population, next
populations
are ordered by similarity to other populations. Clusters (k=8) reveal the ancestral similarities between populations.
B.
Using the pairwise sequentially Markovian
coalescent method, effective
population size (Ne) was determined for each population
for the last 600 thousand years. All humans
shared a demographic history up to about 300 thousand years ago (kya). About
150,000 years ago, non-African
populations experienced a drastic decrease in population size (a
bottleneck).
African populations also experienced a similar long term bottle neck, but
the African
effective population size remained larger than that of non-Africans. In
the
last 60,000 years, most populations have increased in size. The
Bengali in Bangladesh population has
experienced the greatest increase in population size.
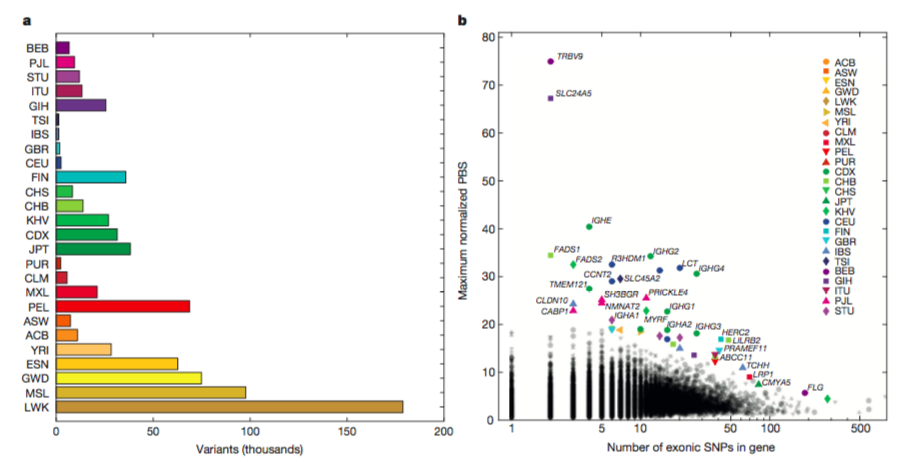
Figure 3.
A. Variants
value on x-axis represents the number
of globally rare variants (frequency <0.5%) that are common
(frequency>5%)
within a population. The Luhya in Webuye, Kenya (LWK) population had the
greatest rare variant number, and populations with European ancestry (TSI,
IBS,
GBR, CEU) had a smaller variant values. Exceptions within continents, such
as
higher than average European variation in the Finnish in Finland (FIN)
population
and lower than average variance for People with African Ancestry in
Southwest
USA (ASW). These findings suggest
that a
portion of rare variation is exclusive to a single population and not to
the
continent and may be indicative of drifted variants.
B.
To identify targets of recent localized
adaptation, FST- based population branch statistic (PBS)
was used. Y-axis
represents maximum PBS value which is indicative genes with strong
differentiation between populations in the same continent. X –axis
represents
the maximum number of exonic SNPs in a given gene. Interestingly, some
of the one
of the most differentiated genes between populations in the same
continent include
TRBV9 (T-cell receptor) and SLC24A5 which is associated with skin
pigmentation. Out of all
variants in each population,
a shockingly low number of genes exclusively differentiated within a
population.

Figure 4.
A.
To determine
if phase 3 data could aid in inferring unobserved genotypes based on human
haplotypes,
9 to 10 individuals from 6 populations were excluded from a reference panel.
Researchers imputed genotypes. The correlation between experimental (omitted
individuals) and imputed
genotypes was determined. As allele frequency increased within a continent,
the
correlation between experimental and imputed also increased a majority of
the
time. Phase 3 data can predict
genotypes
of continental high frequency alternative alleles.
(Bottom
left) Due to increased genotype and sequence
quality, phase 3 data can better correlate experimental (omitted
individuals)
and imputed genotypes in all samples and in intersecting samples as long
as alleles
have high continental frequency.
B. To Determine the average number of tagging variants
(individual SNP that represents a larger group of SNPS) needed for common
(top),
low frequency (middle) or rare (bottom) individual variants in a
population.
African populations have the lowest number of tagging variants for both
common and
low frequency variants. In rare variants, Americans and Europeans have the
highest number of tagging variants, but across all continental groups,
there is
at most a 3 tagging variant difference.
C.
To determine if fine-mapping genetic
association
signals could be derived, expression quantitative trait loci (eQTL) was
used on
69 samples of 6 populations. Percent of indels (darkest color), tied
(medium
color), and SNPs (light color) are depicted.
D.
Populations were combined and a metadata
approach was utilized to determine the percentage of eQTLs in TFBS.
Reference:
The
1000 Genomes Project Consortium. 2015. A global reference for human
genetic variation. Nature 526:68-74. Doi:10.1038/nature15393
*** Unless otherwise cited, all figures borrowed from 1000
Genomes Project Consortium ***
back
to home page
back to
home page
Genomics
Page
Biology Home Page
Email Questions or Comments: itcuellar@davidson.edu
© Copyright 2018 Department of Biology,
Davidson College, Davidson, NC 28035





