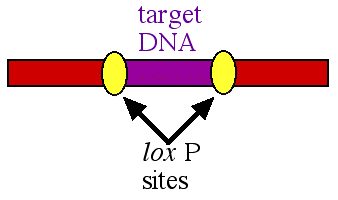
In the early 1990's a new method was developed to delete a specific portion of DNA. The procedure took advantage of the basic research performed on the bacteriophage called P1. In this virus, there is an enzyme called cre and particular DNA sequences called lox P sites. The lox P sites work in pairs and they flank a segment of DNA called a target (figure 1).

Figure 1. A pair of lox P sites (yellow ovals) flanking the target DNA (purple) to be deleted.
When the cre enzyme binds to the lox P sites, it cuts the lox sites in half and then splices together the two halves after the target DNA has been removed (figure 2).
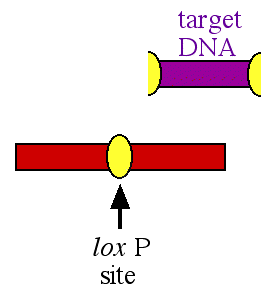
Figure 2. After the cre enzyme has excised the target DNA, one lox P site is left behind and the two flanking fragments of DNA are spliced together. The target DNA is excised and degraded.
Molecular biologists recognized the specificity and utility of this viral recombination system and put it to good use. Now if you want to excise a piece of DNA at a particular time, all you need to do is to flank the target DNA with a pair of lox P sites and introduce the cre enzyme when you want the target excised. Mike Snyder's group used this to added epitope tags onto yeast proteins (Proteomics Chapter).
An additional twist is to express a Cre transgene under control of an inducible promoter so you can delete the target DNA inside selected cells of a transgenic organism when you want it deleted.
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu