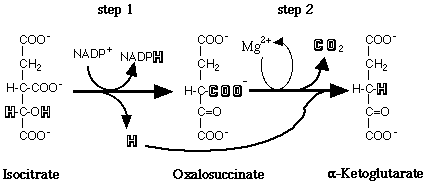

The substrate of the reaction is not free isocitrate, but rather isocitrate accompanied by a divalent metal cofactor (Colman, 1972; Carlier and Pantaloni, 1973). There are two forms of IDH differentiated by their coenzyme requirement and their cellular localization. The NAD+-dependent isocitrate dehydrogenase (E.C. 1.1.1.41) is found in the mitochondria and is present only in eukaryotes. The NADP+-dependent isocitrate dehydrogenase (E.C. 1.1.1.42), also referred to as cytosolic IDH, is ubiquitous in its distribution and is believed to be the prevalent species in most organisms (Chen and Gadal, 1990). This form of the enzyme is considered to be the only type present in bacteria and cyanobacteria. Additionally, NADP+-dependent isocitrate dehydrogenase appears to be present in all organs and tissues in higher plants and animals (Chen and Gadal, 1990). The major form of NADP+-dependent IDH, a homodimer of less than 100 kDa with subunits of 40-50 kDa, is found in most prokaryotes and eukaryotes. A less widely distributed form of NADP+-dependent IDH has been described as a monomer of 55-80 kDa and has been reported in a single bacterium, a fungus, and some mammalian tissues (Chen and Gadal, 1990).
Initial descriptions of NADP+-dependent IDH from Drosophila melanogaster agreed that this enzyme is composed of two subunits. However, Fox, et al., (1971) reported Drosophila IDH to be a dimer of 81 kDa, while Williamson, et al., (1980) estimated the size of the enzyme to be 110 kDa. What was unique about the Williamson, et al., paper was the suggestion that Drosophila IDH is composed of two different subunits, one 50 kDa and one 60 kDa in size.
In addition to the question regarding size and
composition of IDH in D. melanogaster, controversy over the location
of the gene(s) encoding for the enzyme emerged in the late seventies. In
the initial localization of the gene for IDH in Drosophila melanogaster,
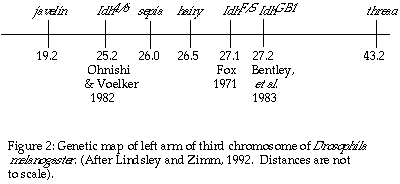
Fox (1971) used electrophoretic variants to localize Idh at 27.1
± 0.4; 0.6 map units to the right of the hairy locus on the
left arm of the third chromosome. This conclusion was accepted until Ohnishi
and Voelker (1979), using electrophoretic variants of IDH, mapped Idh
in Drosophila simulans, a sister species sharing the same banding
pattern on chromosome three with Drosophila melanogaster, to the
left of the sepia locus (26.0). Following through with their suspicion
that the genetic location of the gene for IDH in D. melanogaster
should be the same as in D. simulans, Ohnishi and Voelker (1982)
localized Idh at 25.2 in D. melanogaster, once again to the
left of sepia (Figure 2). The issue was further clouded by the mapping
of a ënullí activity allele, (Idh GB1), to 27.2 in D.
melanogaster by Bentley, et al., in 1983. Attempting to synthesize
the mapping data, Bentley, et al., suggested that the active form
of Drosophila IDH may be encoded by two different genes, that each
gene results in an inactive subunit, and that the active form of IDH is
a dimer of these two gene products.
Our experiment revisited the subunit question
regarding IDH in D. melanogaster, with the specific goal of answering
the "one subunit or two" question. We successfully modified the technique
of Ni, et al., (1987) and purified IDH from D. melanogaster
to apparent homogeneity using ammonium sulfate fractionation and Matrex
Red A affinity chromatography. The concentrated IDH was evaluated for Km
values for both isocitrate and NADP+, divalent metal ion requirements,
and native and subunit molecular sizes.