A Genbank search on the amino acid sequence
of pmh1 revealed that pmh1 has a strong sequence homology
with the plasma membrane H+-ATPase. These plasma membrane H+-ATPases
are found in a variety of organisms ranging from unicellular biflagellates
(T. cruzi, L. donovani etc.) and fungi (S. cerevisiae, K. lactis
etc.) to vascular plants (O. sativa, Z. marina etc.). Multiple sequence
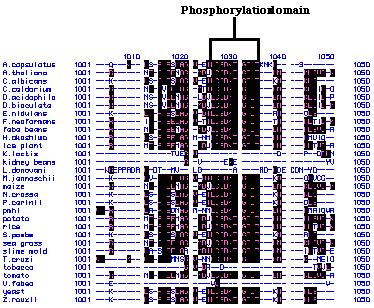
alignments of these peptide sequences showed that there are several highly
conserved regions (Figure 24). For example, the amino acids that code for
phosphorylation sites within these peptide chains are extremely conserved.
A phylogenetic tree (Figure 25) created via MacDNAsis revealed several
expected trends such as the clustering of the unicellular biflagellates,
fungi and vascular plants into their own groups. This shows that the homology
of the peptide sequence in these organisms correspond to the groupís evolutionary
developmental pathway. It is interesting to note that the homology of the
pmh1 sequence in the unicellular biflagellate group is more distantly
related to the other groups than is M. jannaschii, a methanogenic
archeon. Due to the low percentage of relational similarities (9.7%) between
the unicellular biflagellates and the other groups of organisms , it is
plausible that the pmh1 gene codes for a different family of H+-ATPases.
The conserved peptide regions indicate that there are several basic, core
sequences that are needed to code for the H+-ATPases. The remaining
pmh1 sequence that vary from the rest of the organisms code for
the unique portion of the pmh1 H+-ATPase. This
novel H+-ATPase might not be localized in the plasma membrane
even though the Genbank search revealed a strong homology to plasma membrane
H+-ATPases.

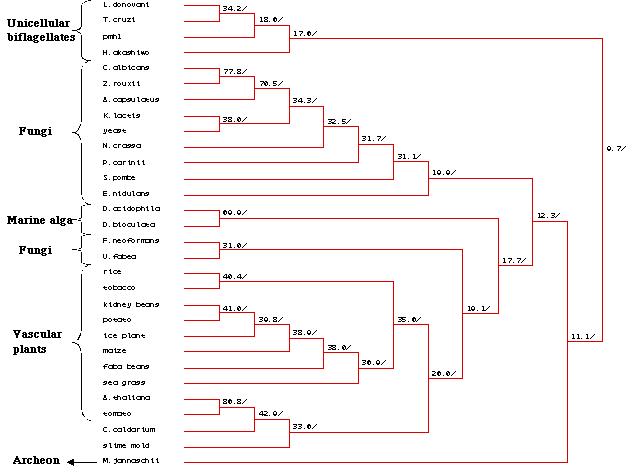
Figure 25: Phylogenetic tree created by MacDNAsis for pmh1 and twenty nine other organisms using the Higgins and Sharp model (Higgins et al., 1988).
Further evidence to support this theory comes
from a Western blot performed on a purified plasma membrane extract from
pmh1 cells (Russo, personal communication). The antibodies used
on this blot were raised against a peptide sequence derived from A.
thaliana plasma membrane H+-ATPases (Serrano, 1993). These
antibodies were capable of crossreacting and binding to C. reinhardtii
plasma membrane H+-ATPases (Norling et al., 1996). However,
the peptide sequence used to raise the antibody cannot be found in the
pmh1 sequence. Thus, the antibody should not be able to recognize
and bind to the pmh1 peptide. Nevertheless, the Western blot of
the purified plasma membrane extract of pmh1 cells done by Kristin
Russo showed that the antibody bound to a protein in pmh1 cells,
possibly a plasma membrane H+-ATPase (Russo, personal communication).
Since the antibody is not supposed to detect the pmh1 sequence,
what is being detected and bound by the antibody is not the pmh1
sequence.
Several models can be proposed to account
for this discrepancy. The Western blot results and the evidence that the
pmh1 sequence is so distantly related to the other groups suggests
that the pmh1 gene codes for a novel H+-ATPase. This
pmh1 H+-ATPase might be localized within the cell and
not on the plasma membrane. In fact, Wada et al. (1994) isolated
and cloned a H+-ATPase with a high homology to plasma membrane
H+-ATPase from Heterosigma akashiwo, a unicellular biflagellate
that is closely related to Chlamydomonas. This H+-ATPase
is expressed in intracellular vesicle-like structures. If pmh1
codes for a novel H+-ATPase, the antibody used in the Western
blot must have bound to the plasma membrane H+-ATPase characterized
by Norling et al. (1996) and not the pmh1 sequence. It is
also possible that the pmh1 H+-ATPase is a new subfamily
of plasma membrane H+-ATPase that is different from the plasma
membrane H+-ATPase characterize by Norling et al. (1996).
In that case, two different types of H+-ATPases might exist
simultaneously on the plasma membrane. If pmh1 codes for H+-ATPase,
further experiments have to be conducted to determine the localization
of the pmh1 H+-ATPase .
In pmh1 cells, the absence of the 4.4 kb message in the RNA blot (Figure 5) indicated a failure of transcription in the pmh1 gene which led to the absence of the H+-ATPase. Without H+-ATPase, the pmh1 mutant strain lacked the capability to transport the H+ cation across its plasma membrane or across membranes of other organelles. Cells regulate intracellular pH mainly through their plasma membrane H+-ATPase (Palmgren, 1998). Thus, if the H+-ATPase was absent or deficient, the H+ cation equilibrium and thus the pH of the cell would be altered. Even if the H+-ATPase is not localized in the plasma membrane, the same argument could be applied to any intra-lumenal compartment within the cell. So, how can we link the disruption of the intracellular pH with the failure of agglutination in pmh1 cells?
The glycosylation of proteins within the Golgi body of a cell is heavily dependent on the intracellular pH. It has been shown that changes in both the extracellular and intra-lumenal pH of a cell "seem to perturb the organization and function of the Golgi" (Varki, 1998). In addition, studies carried out on the Chinese hamster ovary cells showed that a slight pH change severely decreased O-linked glycoslylation in the Golgi (Anderson and Goochee, 1995). Thus, we can surmise that a change in the internal pH of a cell would affect glycosylation of proteins. If our novel pmh1 H+-ATPase is localized in the membrane of the Golgi body or in the trans-Golgi network, disruption of protein glycosylation would be even more severe. Agglutinins are hydroxyproline rich (Cooper et al., 1983) and the O-linked carbohydrate portions represent about half their molecular mass (Samson et al., 1987). Thus, it is very likely that if the glycosylation mechanism within the mutant pmh1 cell is disrupted, then non-functional agglutinins are being made and agglutination fails to occur. Studies have shown that tunicamycin, an N-glycosylation inhibitor, prevented agglutination and gametogenesis in Chlamydomonas (Ray et al., 1982). O-glycosylation can be affected by tunicamycin since drugs that effect N-glycosylation have been reported to alter O-glycosylation patterns (Niam, 1994). The carbohydate portion of agglutinin plays a central role in mating since results have indicated that mild periodate treatment (Samson et al.,1987 ; Adair, 1985) or alkaline borohydrate reduction (Collin-Osdoby et al., 1984) to remove the carbohydate moiety resulted in the complete inactivation of agglutinins.
To corroborate our hypothesis that the pmh1
gene codes for a H+-ATPase, several mutant phenotypes observed
for pmh1 can be attributed to the absence of the H+-ATPase.
(1) A wild type Chlamydomonas has an elongated shape (Figure
26A). However in pmh1, it was observed that all the cells were spherical
(Russo, personal communication) (Figure 26B). The major Chlamydomonas
cell wall constituents are O-linked glycoproteins and not cellulose (Harris,
1989). Both cell walls and agglutinins contain hydroxyproline and serine-rich.glycoproteins.
Thus, if glycosylation is disrupted, the altered cell wall structure will
lose its rigidity and hence, the mutant phenotype (spherical shape of the
cell) is observed.


(2) It was observed that pmh1 grew slower compared to the wild type cells on a regular TAP (Tris-Alkaline-Phosphate) plate. Since internal alkalination and regulation of pH is required to initiate DNA synthesis and cell division in fungi and animal cells,(Palmgren, 1998), a lack of pH regulation in the cell could retard growth.
The evidence presented in this paper has led us to propose that an altered pH environment within the cell, whether it is cytoplasmic or intra-lumenal, has the potential to disrupt the glycosylation process. Since agglutinins are glycoproteins, failure of agglutination in pmh1 cells are caused by the non-functioning agglutinins.
In order to confirm our hypothesis, further molecular characterization of pmh1 needs to done. First of all, transformation of constructs A and B has to be carried out. If complementation is successful, it will allow us to ascertain the identity of the pmh1 gene as the source for the mutation. Once the identity of the mutation is elucidated, functional assays can be conducted to determine whether pmh1 truly codes for H+-ATPase and whether the H+-ATPase is localized in the plasma membrane. Antibodies can be raised against a selected pmh1 peptide so that immunolocalization can be conducted to determine the presence of the H+-ATPase. Alternatively, differential centrifugation and two-phase partioning in an aqueous polymer system (Norling et al., 1996) will allow us to isolate purified plasma membrane H+-ATPase. Activity assays or an immunoblot with antibodies can then be carried out to determine whether pmh1 contains plasma membrane H+-ATPase. Comparison of the pH in intracellular compartments can also be done using flourescein tagged molecules or acridine orange and neutral red dyes. All these reagents are sensitive toward pH. A new method known as PEDAC (Post-Embeding Dectection of Acidic Compartments) can also be used to monitor the H+ gradient in the cell (Anderson et al., 1988). PEDAC uses DAMP (3-[2,4,-dinitroanilino]3íamino-N-methyldipropylamine), a specially formulated weak base that gets accumulated in intra-lumenal compartments proportional to H+ concentrations. A suitable monoclonal antibody can then be utilized to detect DAMP.
Elucidating the function of the pmh1
gene will allow us to understand the importance of intracellular pH regulation
in the glycosylation mechanism. Ultimately, knowledge of how a cell regulates
intracellular pH could have biomedical applications. It has been
shown that pH affects the survival and growth of pathogenic bacteria such
as Heliobacter pylori (Sjostrom et al., 1996). Benzimidazole,
a H+-ATPase inhibitor, has been shown to exert a specific
antibacterial activity against H. pylori (Sjostrom et al.,
1997). In addition, if pmh1 does indeed code for H+-ATPase,
we can discover other cellular processes that are sensitive toward the
H+ cation equilibrium.