Yeast Knockouts - Barcode Approach
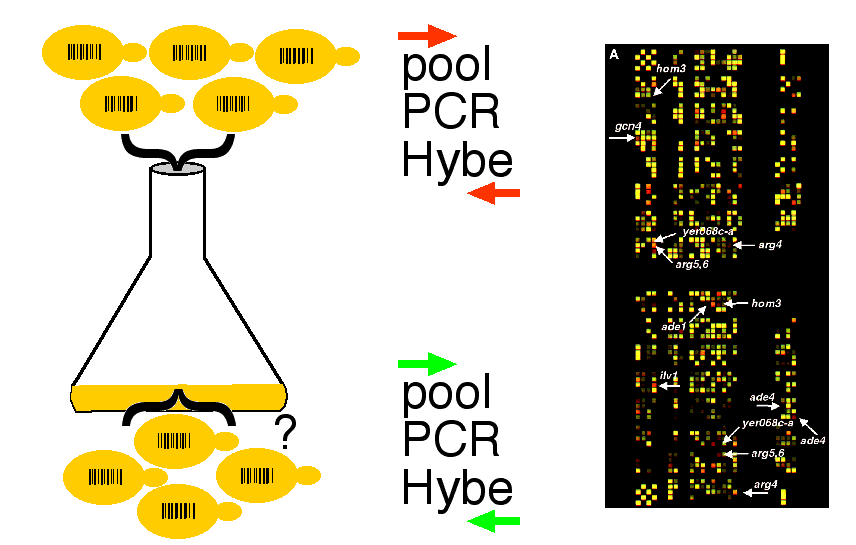
- Delete each gene one at a time and leave a barcode tag (20mer UPTAG and 20mer DOWNTAG) that creates a unique code for each deletion strain.
- Every gene to be targeted, though some are essential.
- Ron Davis et al., (Stanford) created 2026 heterozygous diploid deletion strains.
- 1620 haploid strains viable.
Experiment:
- Mix 500 barcoded mutants into one flask and grow under some environmental pressure (carbon source, temperature, pH, inhibitory drugs, etc.) and let the cells grow.
- Those that lack drug target gene or contain a necessary gene will grow.
- Those that retained drug target or lack necessary gene will not grow as fast.
- Aliquots removed over time.
- Barcodes amplified en masse by PCR and hybridized with microarray.
- Monitor loss of barcoded mutants over time.
- Determine essential genes under selective pressures, drug targets, etc.
Mircoarray Data -
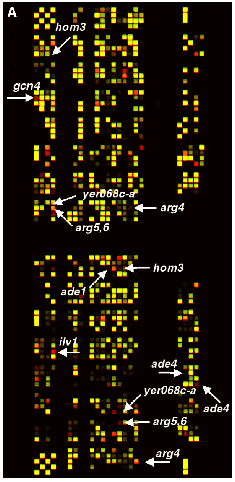
Red label = strains grown for 0 hours
Green label = strains grown for 6 hours
Red spots indicate stains that did not compete well against others.
Graphical Data
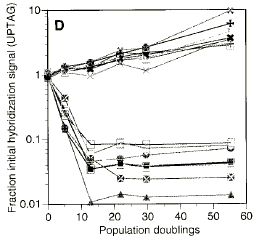
Significant Conclusions for GCAT:
- Discover function of unknown ORFs
- Discover unsuspected functions for known ORFs
- Discover drug targets (direct and indirect)
- 15,300 strains available for further study.
-
- Uncertain correlation between gene expression microarray data and selection data.
- Many strains and an infinite number of environmental conditions.
Caveats
- Cells contain selectable marker neomycin resistance.
- Composition of pooled mutants may be important (secreted complementation).
GCAT
Home Page
Biology Home Page


© Copyright 2001 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu


