Yeast Knockouts - mTn Approach
Michael Snyder et al. (Yale)

Random insertional mutagenesis into yeast gDNA library.
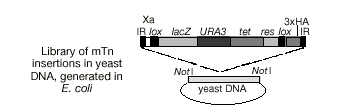
Key mTn features:
- Met- lacZ - creates fusion protein that can be detected with Xgal (no IPTG)
- URA3 - selection in yeast for presence of mTn
- tet - selection in E. coli for plasmid
- two lox sites - allows for Cre-mediated deletion of insertion
- 3xHA - three copies of the haemagglutinin epitope tag to which a monoclonal antibody binds, used to follow your favorite protein after Cre-lox excision.
What's Available?
- 11,232 stains created
- 6,358 strains verified by sequence
- 5,442 in or very near annotated ORFs
- 1,917 ORFs affected
- 1,826 strains have high ß-gal expression as fusion proteins
- 328 non-annotated ORFs (NORFs) discovered (due to small size or overlapping larger ORFs)
Different Levels of Analysis
1) Macroarrays
- Wild-type Expression Patterns "enhancer trap-like" (ß-gal)
- Specific Growth Conditions:
|
Oxidative Phosphorylation
|
Cell Wall Synthesis
|
|
ade2 and red pigment formation
|
lysis and release alkaline phosphatase
|

|

|
|
Red/White
|
Blue/White
|
2) Microarrays
3) Proteomics
- epitope tag protein detection
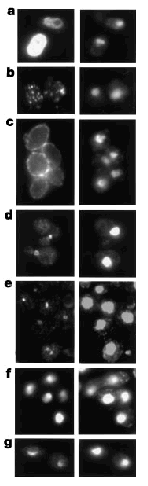
4) "Phenomics"
- Dissection of individual genes since more than one insertion per ORF
(e.g. IMP2' transcription factor with 11 different insertion sites)
- Growth of individual stains on selective media

5) Clustering of Phenomics
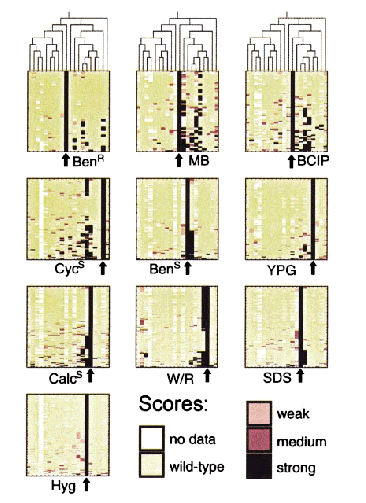
Significant Conclusions for GCAT:
- Discover NORFs.
- Discover function of unknown ORFs.
- Discover unsuspected functions for known ORFs.
- Discover drug targets (direct and indirect).
- 11,232 strains available for further study.
-
- Little correlation between gene expression microarray data and selection data.
- Many strains and an infinite number of environmental conditions.
- Domain mapping with multiple insertions.
- ß-gal fusion provides gene activation method.
- ade2 oxphos detection.
- Macroarray phenomics (96 well or on solid media).
- Proteomics with HA epitope tagging.
- Web-based TRIPLES database for 7800+ mutant strains and online ordering form for strains
(Transposon-Insertion Phenotypes, Localization and Expression in Saccharomyces)
Caveats
- Not every ORF hit due to "random" mTn insertion.
GCAT
Home Page
Biology Home Page


© Copyright 2000 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu






