Experimental Design and Controls
Experimental Design Questions
Define probe v. target
Label RNA v. cDNA (impact on spotted material)
Cy series v. Alexa series dyes
Spots made of oligos v. cDNA
Affymetric chips v. printed glass chips
Reproducibility within and between labs
Dye reversal
Pooled v. control reference mRNA
Control spots from another species
Our Research
How do you know if what you are measuring accurately represents
your sample?
-
Produced 10 PCR products from yeast genome
and 1 from fly genome
-
PCR products matched for GC content and length
-
Made 10 probe pairs; 2 for each gene (two
colors/gene)
-
Each probe matched for GC content and length
-
Probes calculated not to bind to wrong genes
- Spotted range of concentrations for each gene
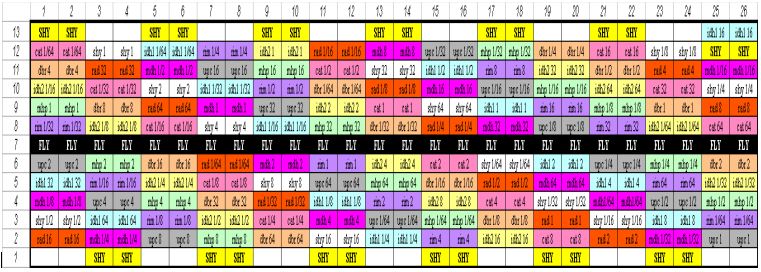
Layout of our DNA Microarray
Calibrated Acquisition of Fluorescence
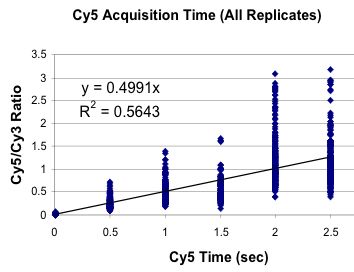
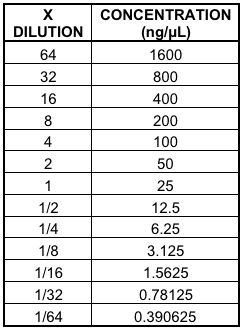
Tested Concentration of Spotted Materials
(4,102 fold range)
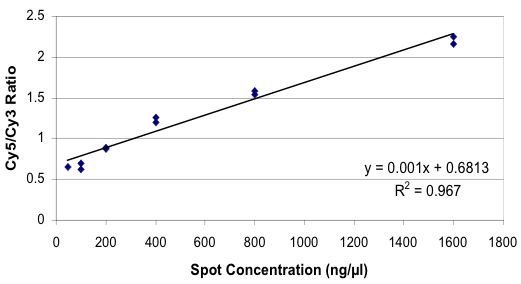
Unexpected Gene-specific Variation
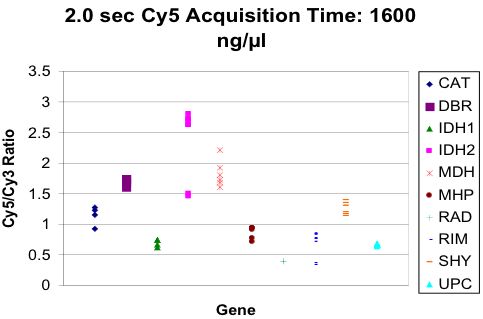
Compared Three Detection Variations
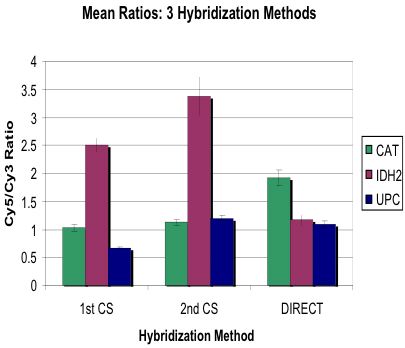
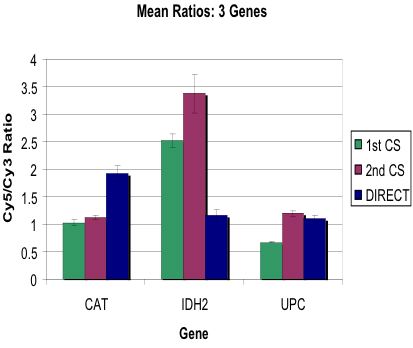
Wanted to Improve Quality of Signal:
Clone PCR Products
Original PCR Products |
Cloned PCR Products |
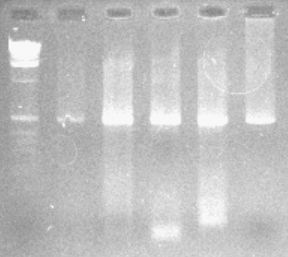 |
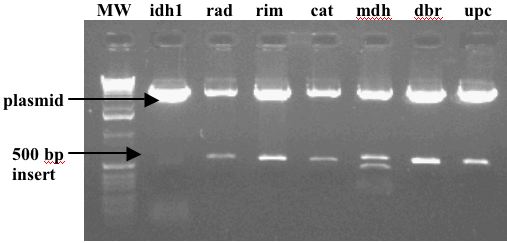 |
Optimized Experimental Conditions:
Signal Up and Noise Down
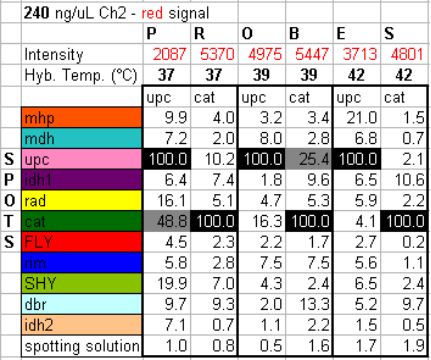
What about Other Ratios? 10:1, 3:1,
1:1, 1:3, 1:10

Good Trends
-
Reproduce each experiment at least 3 times
for each dye (total of 6 times).
-
Spot oligos if possible
to minimize unintentional cDNA-to-spot binding.
-
Verify a few genes (how many?) by alternative
methods (real-time PCR, Northern blots, RNase protection assay, etc.).
-
Use DNA microarrays to detect candidate genes
and overall trends. Do NOT use them to quantify
activity of individual genes.
CSU Workshop
Genomics
Course Page
Biology Department Main Page


Send comments, questions, and suggestions to: macampbell@davidson.edu
or (704) 894 - 2692
© Copyright 2003 Department
of Biology, Davidson College, Davidson, NC 28035












