SNPs (pronounced "snips") are what we used to call point mutations. They are base pair differences among individual chromosomes. Here is an example of a SNP on chromosome B:
chomosome
A: GCATCGTACATGACAGT
chomosome B: GCATCGTgCATGACAGT
chomosome C: GCATCGTACATGACAGT
chomosome D: GCATCGTACATGACAGT
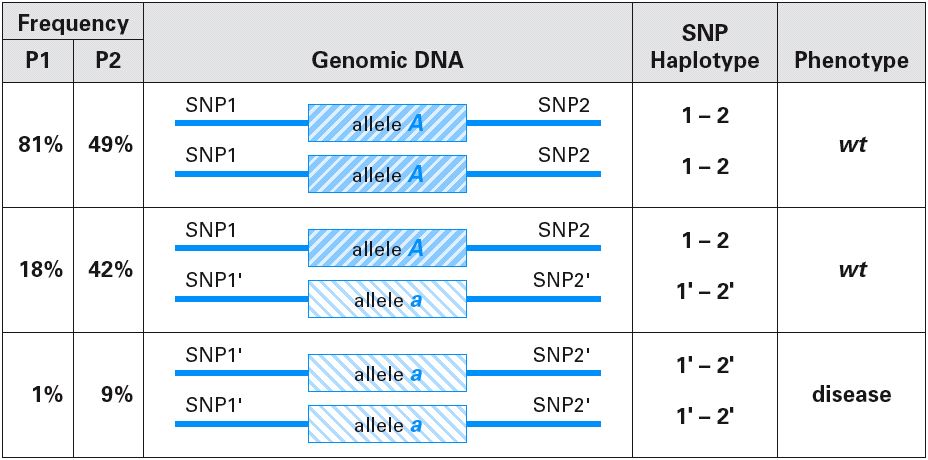
SNPs can be mapped all over the genome and used to map disease loci:

The newest fad in human genomics is the HapMap (Haplotype Map). Haplotypes are a collection of sequences (alleles, SNPs, etc.) that are located close together on the same chromosome.
Recently, it was discovered that humans exhibit a limited number of haplotypes (about 4) at any given location in the genome. If all haplotypes could be mapped, then some people thing polygenic traits could be mapped as well.
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28035