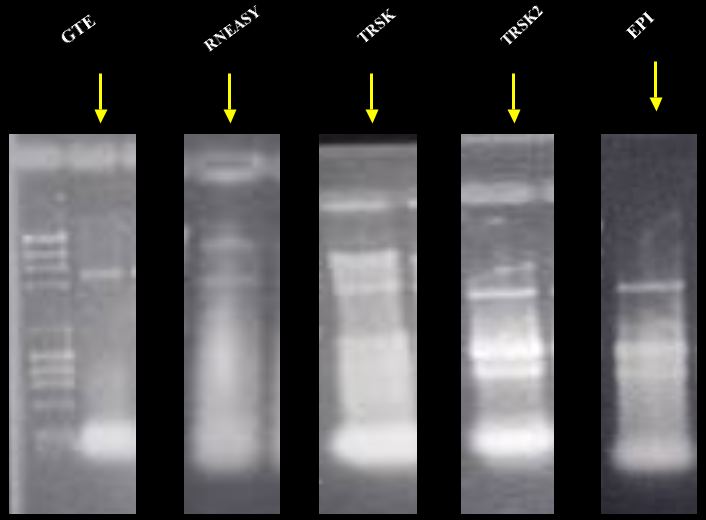
Non-denaturing 0.8 %
agarose gel using DNA ladder
(23, 9.4, 6.6,
4.4, 2.3, 2, 1..4, 1, 0.9, 0.6 Kbp respectively).
Five different gels run about the same amount of time.
* GTE, Guanidine Thiocyanate with later phenol and chloroform extraction.
* RNEasy from Qiagen.
* TRSK, Total RNA Safety Kit with cells OD600 1.5 to 2.0.
* TRSK2, Total RNA Safety Kit with cells OD600< 0.9.
* EP1, Epicentre (Master Pure Yeast RNA Purification Kit).
With my research student, we have tried in three different instances of this academic year the microarray experiment:1) In December 2004, we used the Genisphere 3DNA array 350 kit under SDS hybridization buffer conditions and a TRSK RNA prep. We did not get any visible spots.
2) In February 2005, we used a new Genisphere 3DNA array 350 kit under SDS hybridization buffer conditions and a TRSK2 RNA prep. Again, we did not get any visible spots.
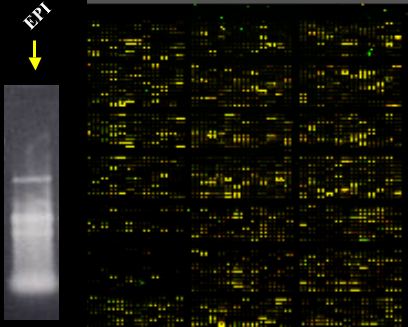
3) In May 2005, we used the same Genisphere 3DNA array 350 kit (from #2 above) under “formamide hybridization buffer conditions" and an EP1 RNA prep. We got great results with the 2 chips we worked during this same day.
Also on the same day as the above Genisphere chips, we used the ISB protocol under “formamide hybridization buffer conditions" and another EP1 RNA prep. We got good results, not as great as with Genisphere, but fine ones. We used only 1 chip (this method requires 50ug RNA).
Very important to mention is that we used the same RT reagents for both protocols because our focus was on the RNA prep quality and we want to minimize the variables. We literally worked in the dark, the lights of the lab were OFF and we managed to follow the instructions with a side light on the corner of the room.
My take home message is that, so far in our hands, the microarray experiment worked when we used RNA prep from EP1 and formamide hybridization conditions regardless of the label production protocol. You probably noticed that we were a little crazy working with three chips and two different procedures on the same day.
My wish is that these short notes will help anyone in need of hope and positive feedback for the microarray experiments. I will be happy to answer any particular question if you write to me.
Consuelo Alvarez, Ph. D.
Assistant Professor of Biochemistry
Longwood University
201 High Street, Farmville, VA 23909
Phone: (434) 395-2847
Fax: (434) 395-2652
E-mail: alvarezcj@longwood.edu