The double-stranded plasmid DNA is independent of the bacterial
chromosomes. Replication of the plasmid chromosomes occurs separately, yet
in the same manner as the bacterial chromosomes (Purves 2001).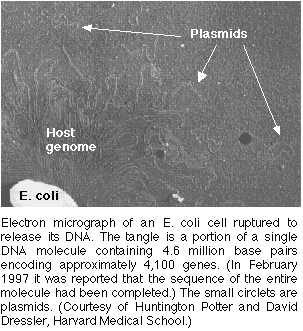
Figures 1&2. Plasmids:Electron Micrograph photo of a plasmid for orientation of size (figure 1) & Permission Pending from Surface Science (figure 2).
The Role of Plasmids
The basic procedure in a gene cloning experiment consists of placing a foreign gene into bacterial cells, isolating the individual cells and growing colonies from each of them. All of the cells in each colony are identical to one another and will contain the foreign gene. Therefore as long as the foreign gene can replicate, the gene can be cloned through the process of cloning its bacterial host. This host serves as a vector, or carrier molecule , for the gene of interest.
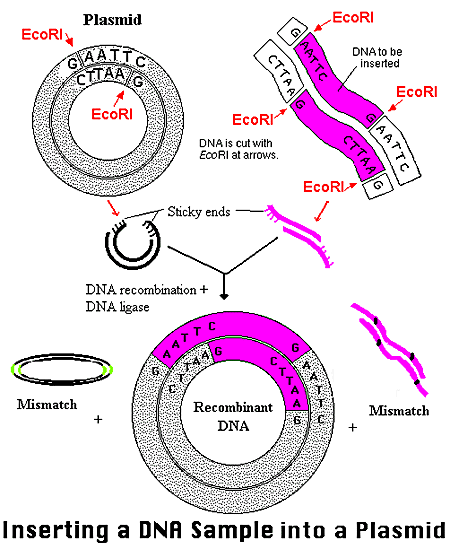
Vectors are divided into two groups: phages and plasmids. Plasmids frequently serve as carrier molecules in gene cloning experiments (Example of Plasmid Use). Foreign DNA in such experiments rely on the plasmid as a carrier molecule for its replication because the gene alone does not contain an origin of replication (Weaver 2002). All of the plasmids contain an origin of replication, but that is not to say that a given gene can be inserted into just any plasmid. The piece of DNA can be inserted as long as both the gene and the plasmid contain recognition sites conferring for the same restriction endonucleases (in nonessential regions). The plasmid and the foreign DNA can then be cut with a particular restriction enzyme (EcoRI in the image below). The result is intermediates with sticky and complimentary ends. The foreign gene and plasmid can then be combined via base-pairing and linked under incubation with DNA ligase. As the sticky ends on the vector and on the gene base-pair momentarily, the ligase acts to seal the nicks, attaching the two DNAs through covalent bonds. The foreign gene is thereby inserted into the plasmid and will remain in place, unless recut with the same restriction enzyme (Roberts 2000).
Figure3. The plasmid and foreign DNA are cut by the restriction enzyme EcoRI in this example. Intermediates are formed with sticky and complimentary ends. With the addition of DNA ligase and incubation, the gene and plasmid recombine by base-pairing at the sticky end sites. The result is a new plasmid containing the inserted foreign gene. A few mismatches do occur in this image, denoting an undesirable recombinant product. *Permission From the National Health Museum To Use This Image Is Pending Approval*
From Micro To Macro: Cloning With Plasmids
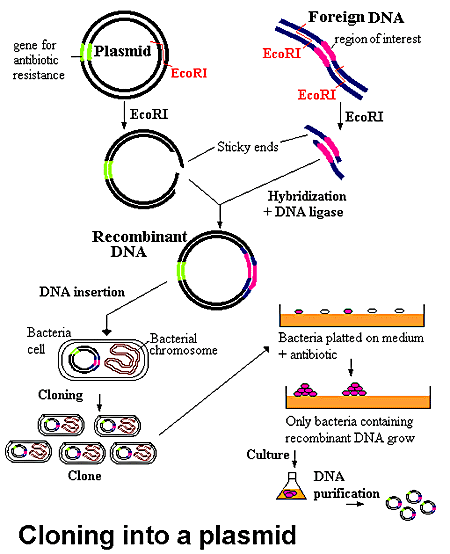
Many diseases are caused by genetic alterations. DNA cloning (enter plasmids) has provided much needed information to enhance our understanding of genetic diseases. In DNA cloning, a DNA fragment that contains a gene of interest is inserted into a plasmid. The plasmid carrying genes for antibiotic resistance, and a DNA strand, which contains the gene of interest, are both cut with the same restriction enzyme (as discussed above). The plasmid is opened up and the specific gene of interest, previously spliced out of its parent DNA strand, is inserted. Upon the introduction of DNA ligase, the gene and plasmid covalently link to form recombinant DNA. The plasmids and copies of the DNA fragment produce large quantities (lots of clones) of recombinant DNA.
At this point it would be simple if all the cut DNA had been ligated to plasmids to form recombinant DNAs, but that never happens (Lin 1984). Rather, the initial clones are a mixture of re-ligated plasmids and re-ligated inserts (genes), as well as the recombinants. This is where the antibiotic resistance genes of the plasmid come into play.
The objective at this point is to grow only bacteria that solely contain the recombinant DNA. Thus the bacteria can be grown in the presence of a particular antibody for which the plasmid is resistant. This type of growth selects for cells that have taken up either the plasmid alone or the plasmid plus the inserted DNA. Cells that do not receive DNA or received only inserted DNA will not be resistant to the antibody; these cells should fail to grow. Narrowing the selection procees down even more, the next task is to find the clones that have received recombinant DNA (Weaver 2002).
Figure 4. The recombinant DNA is allowed to transform a bacterial culture and then plated in the presence of antibiotics. All the cells which have been encoded by the plasmid DNA recombinant are killed. The remaining cells contain the desired recombinant DNA. *Permission from the National Health Museum To Use This Image is Pending Approval*
The Basics of Phages
![]()
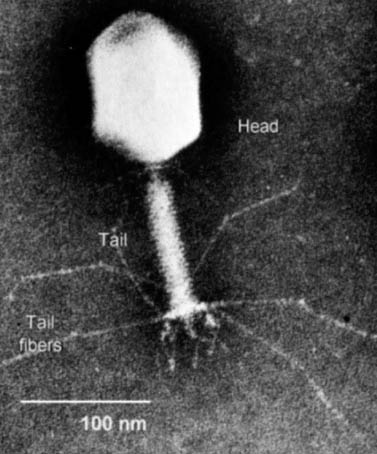
Figure 5. Electronmicroscope Photo Image of Bacteriaphages. This is just a basic photo of phage vectors to provide a general idea of how phages appear. Permission To Use Image Is Pending Approval.
Phages are another group of natural vectors. Bacteriophages transduce bacterial DNA from one cell to another (Purves 2001). Phage vectors have a natural advantage over plasmids: they infect cells much more efficiently than plasmids transform cells, so the quantity of clones yielded with phage vectors is typically greater. Using phages, clones are not colonies of cells, but plaques formed when a phage clears out a hole in a lawn of bacterial growth. Each plaque of derives from a single phage that infects the cell, killing it and the cells in vicinity (Ausubel et al 1992). The degradation process continues until a visible patch, or plaque, of dead cells appears. Since these phages in the plaque derive from one original phage, they are all genetic reprints of one another, or clones.
Bacteriophages, or phages for short, are viruses whose hosts are bacterial cells. Like all viruses, phages are metabolically inert in their extracellular form, and they reproduce by insinuating themselves into the metabolism of the host. The mechanisms by which phage virions infect their host cells vary among the different types of phages, but they all result in delivery of the phage genome into the cytoplasm of the bacterial host, where it interacts with the cellular machinery to carry the phage life cycle forward. The result of infection can be, and often is, total devastation for the cell. A good example of this is infection by the E. coli phage T4, which commandeers the material and energetic resources of the cell and turns them toward making more virions, after which it causes violent lysis of the cell and release of the progeny virions. At another extreme, the large group of phages known as temperate phages have the option when they infect of setting up a state of coexistence with the host (“lysogeny”) in which the genes that would harm the host are prevented from being expressed, while a small set of genes that provide benefit to the host are expressed. Both scenarios result in replication and perpetuation of the bacteriophage (Purves 2001).
Comprehension of the intricate nature of phages has been used, like the types just explained, in laboratory scenarios as one of the many different molecular tools within cloning processes.
Figure 6.Electron Micrograph of a Bacteriophage for general structural or orientation purposes. Not the sizes of the head, tail, and tail fibers. Permission to Use this Image is Pending.
Focusing On A Specific Tool: pBluescript
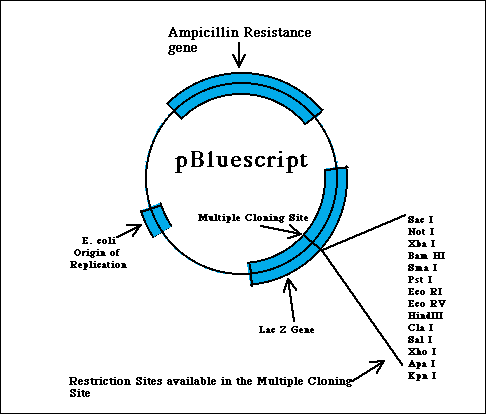
pBluescript is an example of a combination between plasmids and phages, thereby gaining it a place within the catergory of vectors known as phagemids. Phagemids represent a hibrid type of class of vectors that serve to produce single-stranded DNA. pBluescript is a popular variety of phagemid commercially available to molecular biology labs for cloning purposes. Through the following descriptions of some key parts of pBluescript, characteristics of plasmids and phages (as discussed above) should be evident (Weaver 2002).
Some useful features of pBluescript: Like pUC vectors, which pBluescript is derived from, there is a multiple cloning site inserted into the LacZ' gene, so clones with inserts can be distinguished as blue versus white cells by staining using X-gal. pBluescript also contains the origin of replication of the single-stranded phage f1, which is related to M13 phage vectors. This translates into that a cell harboring a recombinant phagemid, if infected by f1 helper phage that supplies the single-stranded phage DNA replication components, it will produce and package single-stranded phagemid DNA. A final feature of this class of phagemid vectors stems from the fact that it is flanked by two different phage RNA polymerase promoters. More specifically, pBS has a T3 promoter on one end side and a T7 promoter on the other terminal end. This is key because it enables one to isolate the double-stranded phagemid DNA and transcribe it in vitro with either of these two phage polymerases to produce pure RNA transcripts to coincide with either of the two different strands (Brunel 1998).
Key Points: