*This page was produced as part of an undergraduate course at Davidson College*
My Favorite Yeast Protein: Proteomics
In previous assignments, I have identified and analyzed genomic data from Saccharomyces cerevisae (yeast) for both an annotated and an un-annotated gene (Assignment One). I have also analyzed expression data from microarrays involving my chosen genes (Assignment Two). This assignment will attempt to make better light of the proteome revolving around these genes through the use of both databases and possible experiments derived from the field of proteomics. It is with a greater understanding of the proteome that I hope to further validate or alter my predictions from earlier investigations. I will first present data regarding the proteins involved with my annotated and un-annotated genes and lastly I will propose possible experiments that could be conducted along with possible results.
FUR4- The Annotated Gene
FUR4, a gene whose function is to produce the enzyme uracil permease, is a member of a family of genes that is located on chromosome 2 in Saccharomyces cerevisae. Uracil permease is a symporter within the plasma membrane that functions by generating an electrochemical gradient across the membrane that is permeable to uracil. It is very similar to other types of integral membrane proteins in that it has a hydrophobic core with structure that is largely determined by the presence of three charged residues in the transmembrane domain.
I decided that one of the first logical steps that I could take would be to attempt to discover an overall structural model of the FUR4 protein. Thus, I attempted a search of PDB, the Protein Data Bank (PDB; 2003, <http://www.rcsb.org/pdb>). Unfortunately, there are no 3D structures available for the FUR4 protein.
The second step that I took was to make use of a database known as the Triples Database (Triples Database; 2003, <http://ygac.med.yale.edu/triples/basic_search.asp>). This is a database in which series of experiments were conducted where transposons were used to disrupt genes in order to produce mutant phenotypes. These phenotypes, due to alterations in protein expression from the gene in question, would give greater insight as to the causative proteins functioning. The following results were obtained:

Figure 1- Results of the Triples Database search (Triples Database; 2003, <http://ygac.med.yale.edu/triples/basic_search.asp>)
There have been several different transposon insertion experiments. In each of these cases it is noted that the expression levels (in the lacZ coloring trials) were for the most part equal during both vegetation and sporulation with low to moderate levels of protein being seen. The phenotypic disruption data available for the final entry on these search results stated this:

Figure 2- Phenotype Disruption Data from the Triples Database (Triples Database; 2003, <http://ygac.med.yale.edu/triples/basic_search.asp>)
There appear to be no significant differences between the wild type yeast and the mutant yeast when FUR4 is altered in this way. I hypothesize then that this could mean that there are other proteins or mechanisms in the cell by which the function of uracil permease is being done without its wild type performance level (isozymes, perhaps?). It could also be that this transposable element did not alter a significant portion of FUR4 and so the product was still largely functional.
Thirdly, I decided to search the Yeast Resource Center's Two-Hybrid Analysis Database (YRC Two-Hybrid Analysis; 2003,<http://depts.washington.edu/%7Eyeastrc/th_11.htm>). If a greater technical understanding of this method is needed it can be viewed here. Basically, if two proteins are found to interact by this method this means that the proteins have bound and have activated the His3 gene which enables the yeast colony to grow on a medium that is His-. Thus, the results here would show what proteins were found to interact with FUR4 in this way. Unfortunately, there was no recorded data for FUR4 upon both a search of the main and supplementary material.
Fourth, I searched the Swiss 2-D Gel Page (Swiss 2-D Gel; 2003, <http://ca.expasy.org/cgi-bin/ch2d-search-ful>. The gels on this website are 2D representations of all the measurable proteins present at a particular time within a cell under particular experimental conditions. Unfortunately, no data was present for FUR4.
I next browsed DIP, the Database of Interacting Proteins (Database of Interacting Proteins; 2003, <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>). This is a site that creates graphical maps of known data to better illustrate networks of interacting proteins. The data obtained in a search for FUR4 is seen below:

Figure 3- Protein Interaction Map for FUR4 from DIP <Database of Interacting Proteins; 2003, <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>)
FUR4 is located in the lower right of this image and it doesn't appear that any known proteins are interacting with it. However, there is a red line connecting FUR4 to ADE2 (the middle node). ADE2 is phosphoribosylaminoimidazole carboxylase, which is thought to be involved in carbon dioxide fixation and purine nucleotide biosynthesis. This is not a confirmed interaction line as denoted by the red color but it is interesting to note that it is at least suspect.
Believing the other databases at my disposal only possible of yielding information that I already had discovered through the use of others, I began to think about the conclusions one could draw from this data and perhaps possible experiments one could conduct to learn more.
Possible Experiments-
1) ADE2-
Since it is suspected but as of yet unverified as to whether or not ADE2 and FUR4 interact as seen above in the DIP diagram, I believe it would be beneficial to confirm this for sure. It might yet provide further insight into the functioning of uracil permease in an auxillary function or perhaps change our understanding of the primary function. There is no data on file for the yeast two-hybrid analysis experiments. It would be interesting to compare FUR4 and ADE2 in this aspect. I believe that these two proteins would enable yeast growth in the His- region. From what I have seen there is more to uracil permease than meets the eye in its functioning (the seemingly unchanged behavior in the TRIPLES evaluation, the DIP diagram, etc.) and it would not shock me to see this gene associated with FUR4.
2) Lox Knockout Experiments-
I am also interested to see more of a wholesale phenotypic effect of the deletion of FUR4 since this type of experiment doesn't seem to be on record. I believe that the construction and proper insertion of an mTn with lox sites that can be activated later on with a viral vector to delete FUR4 completely from the genome (this would be confirmed with further proteomic experimentation such as a 2D gel or protein microarray) would provide great insight as to the phenotypic effects of this protein. If things proceed as I believe they would and the full deletion of the gene would not cause any observable major phenotypic effects, further experiments could be done to gather information on what is expressed in the proteome under this condition to see if increased amounts of any particular proteins are present. This would confirm my previous suspicion as to whether or not there is some other gene/protein at work to compensate for or aid in the functioning of FUR4.
YBLO10C- The Un-Annotated Gene
Going into this assignment, I knew very little as to the nature of YBL010C the hypothetical nearby neighbor to FUR4. From the previous two assignments, two main things had been hypothesized: that this was involved somehow in DNA binding and that it was either located in the cytoplasm or the membrane. Thus, I felt the best thing to do would first be to do a search of MIPS.
MIPS (Comprehensive Yeast Genome Database- Munich Information Center for Protein Sequences; 2003, <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YBL010c>) this comprehensive database shed a great deal of insight and confirmed that there had been very recent discoveries as to the role of this un-annotated gene.

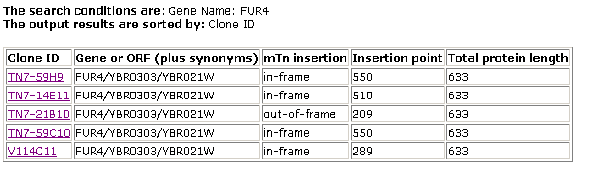
Figure 4- The results of the search of MIPS
The protein has been localized to the cytoplasm. This would appear to have been done in an experiment using fluorescent epitope tags by Kumar et al. in 2002. This confirms my earlier hypothesis as to the cellular localization of the protein. More specifically it was found by Huh et al. in 2003 to be localized and involved with golgi-vacuole transport vesicles. Intrigued by this, I linked using MIPS to SGD (SGD; 2003, <http://db.yeastgenome.org/cgi-bin/SGD/>) to learn more about its functioning in relation to golgi-transport vesicles and observed the following image.
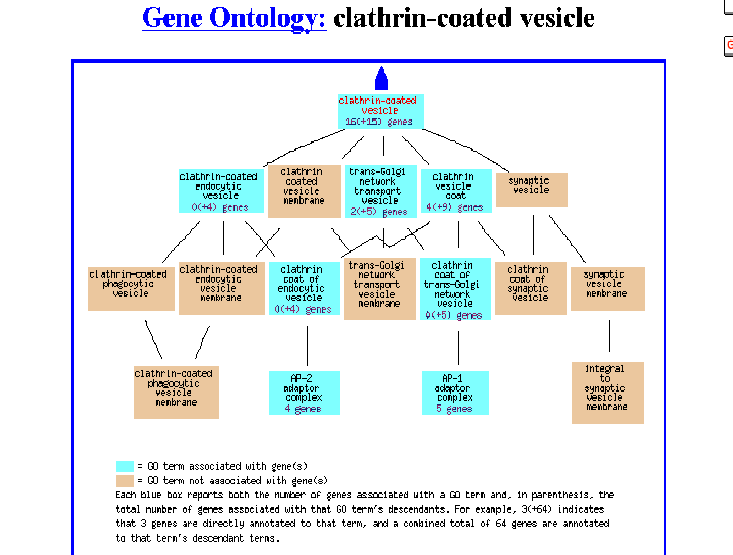
Figure 5- The Gene Ontology of this protein as a clathrin-coated vesicle
Apparently, it was identified as being associated with a clathrin coated vesicle involved with transport in the Golgi. For me, this is indeed expected and refreshing as during the previous two assignments this type of information simply did not exist.
A search of the PDB for a structure of YBL010C again did not yield any results.
Upon searching DIP, I found the following image:
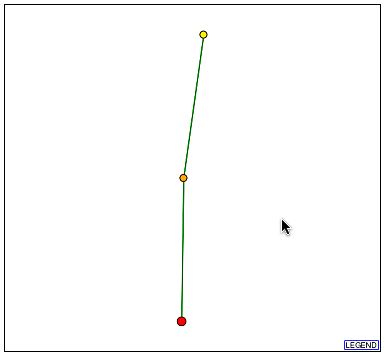
Figure 6- The Protein Interaction Map from DIP for YBL010C
It can be seen that there is one protein with a direct and confirmed connection to YBL010C. This protein, according to DIP, is hypothetical and un-annotated. It is known as YKR022C and although its function is unknown it is apparently vital to cell function. The profile of this protein can be seen here at SGD. Two things are known about this protein: one is that it is localized to the cytoplasm, endoplasmic reticulum, and the nucleus and the second is that in a full-scale deletion experiment the cell phenotype was unviable. This is extremely significant as YBL010C is perhaps involved in the transport of these proteins as it is possibly associated with golgi transport vesicles and was localized to the cytoplasm.
Next, I decided to search the TRIPLES database. The information that I found to be of note is depicted below.

Figure 6- The results of a TRIPLES search (Gene Expression Data) during LacZ testing for YBL010C
It is shown here that during the Vegetative state an intense blue reading was seen. This is significant and gives further insight as to the functioning of the protein. Perhaps during the vegetative state it aids in some sort of metabolic or reproductive function as hypothesized before? If it is related to golgi transport as guessed, perhaps some process during the vegetative state requires for greater transport of particular proteins that the un-annotated gene binds or surrounds? Since a large amount of it is expressed then it is not beyond all consideration for this to be true.
Upon a search of the Swiss 2-D Gel Page, I found that there was no additional information on YBL010C.
Searching the Yeast Resource Center's Two-Hybrid Analysis Database also yielded no results on YBL010C.
Thus upon utilizing these relatively new proteomic tools, we see the further illumination of a topic that was largely in the dark in my previous investigations. My two previous guesses as to the nature of this protein were found to be correct but most unexpected perhaps was its association with golgi transport vesicles. Further research within these veins will no doubt glean more information on YBL010C.
Possible Experiments-
1) What is binding to YBL010C?
An idea for how to go about doing this would be to use GST Pulldown Assays. One could suspend YBL010C protein within a GST matrix and then expose that matrix to cell extract (the proteome) from one of the clathrin-coated vesicles. Afterwards, one could use monoclonal antibodies for desired proteins to probe for suspected proteins (especially YKR022C) and confirm or deny their interaction with YBL010C. I cannot fathom as to what the results of this experiment could possibly be; however, I do imagine that a result would shed further light as to the reason why these vesicles are being shifted around the golgi with the involvement of YBL010C.
2) Where are the vesicles heading?
I believe that some sort of GFP or immunoflourescence modeling with YBL010C as an epitope would be useful in tracking the migration of these vesicles that might utilize YBL010C. The cellular destination of these vesicles and where YBL010C appears to be localized with regard to these vesicles would definitely give insight as to the biological function of this un-annotated gene. Personally, I believe that these vesicles are more than likely confined to near or around the golgi and nucleus due to their previous hydrophobicity ratings and the data would show this. This is even more intriguing due to the information uncovered regarding YKR022C. Observing where the YBL010C vesicles travel could confirm its involvement as a golgi transport protein of the YKR022C protein in general.
References:
Kumar A et al.: Subcellular localization of the yeast proteome. Genes Dev. Mar, 2002; 16(6):707-19.
Email Matt: matalbert@davidson.edu
Return to the Genomics Homepage- click here
Return to the Davidson Biology Homepage- click here
Return to Matt's Genomics Homepage- click here