The gene SSK1 is found on chromosome 7 in Saccharomyces cerevisae. The protein it encodes is part of a two-component osmosensor that regulates a MAP kinase cascade in response to hyperosmotic stress; when activated, SSK1 induces the MAP kinase cascade to raise the level of glycerol in the cell to restore osmotic balance. For more information about this gene, see My Favorite Yeast Genes.
The expression patterns of SSK1 can be analyzed using data from DNA microarray experiments. For an overview of microarray technology, click here. The following DNA microarray results were obtained from the Expression Connection, an online collection of data from multiple DNA microarray experiments (Expression Connection, 2004).

Figure 1: The color scale used to gauge the repression or induction of each gene in the DNA microarray results. More intense colors represent greater gene repression or induction. Black represents no difference in expression between the experimental and control conditions (Expression Connection, 2004).
EXPRESSION OF SSK1 DURING SPORULATION (Chu S, et al)
Sporulation is the process by which yeast cells produce spores after meiosis. As seen in Figure 2, SSK1 (also called YLR006C) appears to be fairly strongly repressed at two hours and remains repressed for the duration of spore production. The yeast cell's size, shape, and internal structure would change quite a bit as the spore forms and matures, so perhaps repressing responses to osmotic stress is a means of conserving energy until the cell resumes normal activity. Other genes with similar expression patterns are also involved in stress response pathways; SSQ1 participates in unfolded protein binding and helps maintain iron homeostasis, and HEX3 responds to DNA damage. There is also the possibility that SSK1 is involved in cytoskeletal organization: SSK1 activates SSK2, a protein whose biological processes include not only the osmosensory pathway but actin cytoskeleton organization and biogenesis, as well. Although there is no DNA microarray data for expression of SSK2 during sporulation, the gene PIN3, which is involved in actin cytoskeleton organization and biogenesis, does exhibit an expression pattern similar to that of SSK1, suggestive of a connection between this molecular function and SSK1 (Expression Connection, 2004).
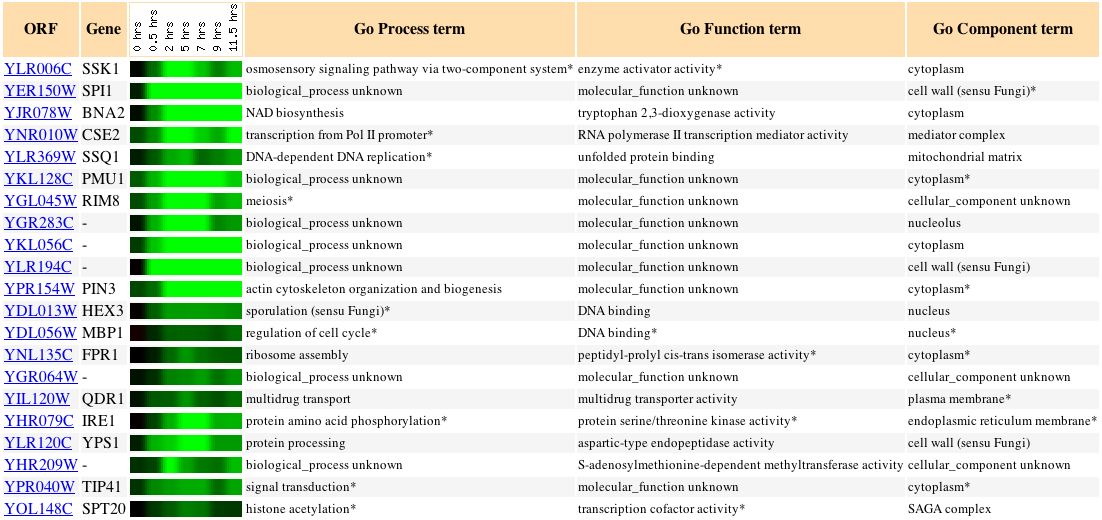
Figure 2:
Expression during sporulation of SSK1 and other yeast genes with a p score of greater than 0.8 (Expression Connection, 2004).
EXPRESSION OF SSK1 DURING THE UNFOLDED PROTEIN RESPONSE (Travers KJ, et al)
The unfolded protein response regulates the expression of genes involved in protein folding, assembly, modification, and degradation in response to stress in the endoplasmic reticulum, i.e. the accumulation of unfolded proteins. As seen in Figure 3, SSK1 is induced during this response; gene induction peaks after two hours. These data suggest that, in addition to the osmosensory pathway, SSK1 plays a role in other stress responses, too (Function Junction, 2004). DNA microarray data from similarly expressed genes can be seen in Figure 4. Although no microarray images of SSK1 expression during the unfolded protein response were available from Expression Connection, it is clear from these data that SSK1 is induced in response to the accumulation of unfolded proteins, and that gene induction is greatest at two hours. The functions of some of the similarly expressed genes are of note with regard to SSK1. The gene NTH1 is involved in trehalose catabolism and is required for recovery from heat shock. It has also been implicated in a response to osmotic stress, which may or may not be a connection to the SSK1 osmosensory pathway. HYM1 is a transcription repressor involved in cytokinesis and the regulation of cell shape; its similarity of expression to SSK1 could indicate that SSK1, "guilty by association," is also involved in cytoskeletal organization. Interestingly, the gene CSE2 appears in the result of this study as well as the sporulation study. CSE2 is required for the regulation of RNA polymerase II; its expression patterns are so similar to those of SSK1 that it might be a transcription factor necessary for the expression of SSK1 (Expression Connection, 2004).

Figure 3: Expression of SSK1 during the unfolded protein response. SSK1 is induced almost two-fold after two hours of exposure to the experimental condition (Function Junction, 2004).

Figure 4: Expression during the unfolded protein response of yeast genes whose p score is greater than 0.8 compared to SSK1. The microarray image for SSK1 was not available (Expression Connection, 2004).
EXPRESSION OF SSK1 DURING THE DIAUXIC SHIFT (DeRisi, et al)
A diauxic shift is a change from anaerobic to aerobic respiration. While the repression of SSK1 in response to this change does not appear to be very strong, as seen in Figure 5, the similar expression of various other genes makes this experiment worth analyzing. The gene UBC4 produces a ubiquitin-conjugating enzyme that is involved in stress responses and sporulation. SPO4, a phospholipase, is also involved in sporulation. CDC3 is required for cytokinesis and acts as a structural component of the cytoskeleton. PIN4 acts as a DNA damage checkpoint during mitosis and becomes hyperphosphorylated in response to DNA damage. The products of several other genes participate in protein transport, targeting, and catabolism. These functions and processes popped up in the two previously mentioned studies, as well, which again implies that SSK1 has molecular functions beyond the osmoregulatory response system (Expression Connection, 2004).

Figure 5: Expression during a diauxic shift of SSK1 and other yeast genes with a p score of greater than 0.8 (Expression Connection, 2004).
EXPRESSION OF SSK1 IN RESPONSE TO DNA-DAMAGING AGENTS (Gasch AP, et al)
Consistent with the similarity of expression between SSK1 and various genes involved in responding to DNA damage, SSK1 expression is affected by exposure to some DNA-damaging agents, as seen in Figure 6. SSK1 is induced in response to heat in the wildtype yeast and two mutant strains, both of which are defective in the Mec1 damage signaling pathway. SSK1 is repressed in response to gamma irradiation in both the wildtype and mutant cells; when exposed to mock irradiation, however, SSK1 is induced in the wildtype and one of the mutant strains after approximately one hour. When exposed to 0.02% MMS (a methylating agent), SSK1 is slightly induced in wildtype yeast, but slighty repressed or unchanged from the control condition in the two mutant strains. These data increase the likelihood of a connection between SSK1 and heat shock and other stress responses; perhaps SSK1 is also involved in the response to DNA damage and is regulated by the Mec1 pathway (Expression Connection, 2004).

Figure 6: Expression of SSK1 in response to various DNA-damaging agents (Expression Connection, 2004).
EXPRESSION OF SSK1 IN RESPONSE TO ENVIRONMENTAL CHANGES (Gasch AP, et al)
This huge experiment looked at yeast cells' responses to many different environmental conditions. From the microarray data seen in Figure 7, it is obvious that SSK1 is involved in multiple stress responses. SSK1 is relatively highly induced in response to severe or prolonged heat shock, hydrogen peroxide, diamide, sorbitol, menadione, and nitrogen depletion (which causes the process of sporulation to begin). SSK1 is relatively repressed or unchanged from the control in response to a drop in temperature, DTT, hypo-osmotic stress, and diauxic shift. These results are consistent with the findings from the previously mentioned experiments, which leads me to conclude that SSK1 is involved in multiple biological processes beyond its known function in the osmosensory pathway (Expression Connection, 2004).



Figure 7: Expression of SSK1 in response to various environmental conditions (Expression Connection, 2004).
CONCLUSIONS
DNA microarray data can be a powerful tool in the exploration of gene expression and function. The gene SSK1 is an annotated gene whose molecular functions, biological processes, and cellular component are already known, but as demonstrated by the above microarrays what we know about SSK1 may be more or less the tip of an iceberg. It is clear that SSK1 is involved in more processes than just the hyperosmotic stress response, or at the very least that SSK1 is co-regulated with genes in other stress response pathways. SSK1 may also be involved in maintaining yeast cells' internal structure and organization. Obviously there is still research to be done; it will be interesting to observe what new discoveries are made in the future concerning the function of this deceptively simple gene.