Figure 1: SynIII design
- Panel A: The S. cerevisiae III chromosome and the synIII chromosome are both shown here. While the III and synIII chromosomes share a repeat as well as non-essential, uncharacterized, and dubious Open Reading Frames (ORFs), the synIII chromosome also has loxPsym sites before non-essential ORF YCR098C and after the repeat, and an added universal telomere cap.
- Panel B: As an example, this shows us how the insertion of a loxPsym site—a palindromic DNA sequence—is inserted into an untranslated region of the 3’ strand, right next to a non-essential gene (in this figure, non-essential ORF YCL055W).
- Panel C: This figure shows the simple change in an essential ORF (YCL004W) from TAG toTAA creates a synthetic stop codon in synIII.
Figure 2: SynIII construction
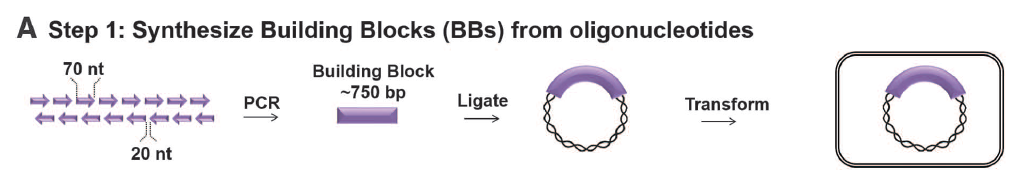
- Panel A: This shows the first step in building synIII—creating the small BB’s from oligonucleotides. These oligonucleotides (60-79 nt’s long) were linked by PCR into BB’s, which could then be ligated into a plasmid that was transformed.
- Panel B: Here we see a depiction of the BB’s from the plasmids and a shuttle vector being combined and cotransformed; from there, recombination occurs so that the BB’s (which contained overlapping pieces of DNA to allow for recombination) merge to form a minichunk of 2-4 kb. “RE” denotes a restriction enzyme cutting site, for either the rare XmaI or NotI, placed on either side of the minichunk.
- Panel C: This shows how the minichunks were used to replace parts of the natural yeast chromosome III, creating synIII. The minichunks, designed to overlap, were recombined such that they merged together, and then into the chromosome. In transformation 1, minichunks 1-4 recombined, with genetic marker LEU2 and a native yeast chromosome III sequence (Linker L1) attached to minichunk 4; Linker L1 allowed the merged minichunks to recombine with chromosome III, producing a hybrid identifiable by the genetic marker LEU2. In yeast transformation 2, similar steps occurred, this time merging minichunks 4-13, with Linker L2 (genetic marker URA3 and a native chromosome III sequence) attached to minichunk 13. Minichunk 4 and Linker L2 allowed for recombination into the hybrid product of transformation 1, producing a synIII section that has replaced parts of native chromosome III with minichunks 1-13 and identifiable by the presence of genetic marker URA3 and absence of LEU2.
Figure 3: Characterization and Testing of the synIII strain
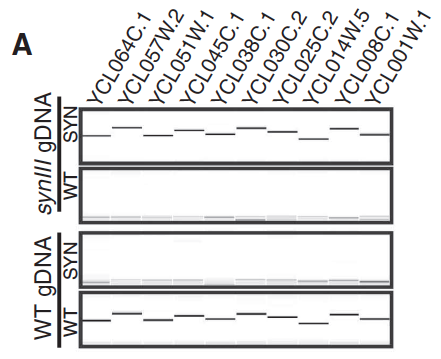
- Panel A: This panel shows us that when the left arm of WT yeast chromosome III was tested, there were matches for every WT PCRTag but no matches for synIII PCRTags; the opposite result was obtained when the left arm of synIII was tested, with no WT matches and all synIII matches.
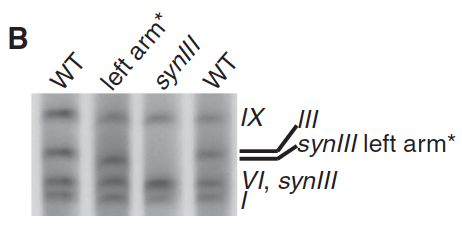
- Panel B: This panel is a gel which has labeled the bands of native III, synIII left arm, synIII, and native IX, VI, and I. In the far right gel lane, we can see that the synIII left arm is slightly smaller than the native III; synIII is even smaller, and is the same size as native VI. The purpose of the “*” on synIII left arm is unclear.
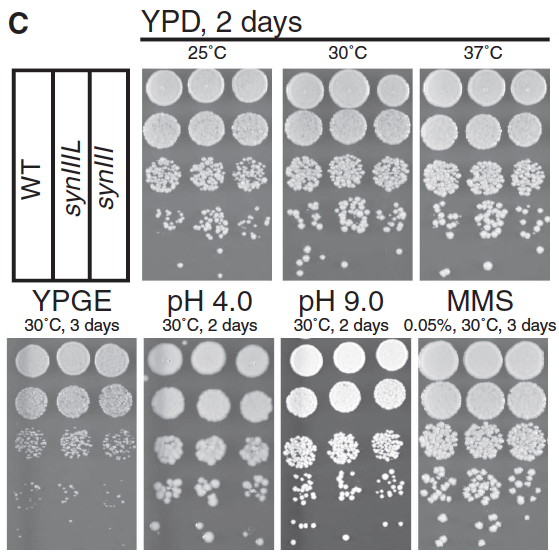
- Panel C: This panel shows us the phenotype and survival of WT, synIIIL, and synIII in serial dilutions on various media and over time. For those grown for 2 days on yeast extract peptone dextrose (YPD), the yeast grown at 25oC look roughly the same no matter their chromosome; synIIIL looks to be perhaps slightly more successful. Similar results hold true for 30oC and 37oC, though colonies are larger across the board as temperature rises. For the rest of the plates (yeast extract peptone glycerol ethanol for 3 days at 30oC, pH of 4.0 or 9.0, both for 2 days at 30oC, and 0.05% methyl methanosulfate for 3 days at 30oC), the general trend seems to be that strains with WT, synIIIL, and synIII are about equal in growth and survival; if anything, the synIII strain is slightly less viable, but synIIIL is comparable to WT.
Figure 4: Genomic stability of the synIII strain
- Panel A: Panel A depicts the methods used to test 30 strains of yeast containing the synIII chromosome for the ability to keep synIII over about 125 generations, a statistic measured by assaying cells for 58 distinct segments lacking essential genes; synIII proved remarkably stable with no deletions occurring.
- Panel B: This panel compares the average loss rate of synIII to the average loss rate of native III; there proved to be no significant difference between the two, indicating that synIII is about equally likely to be lost as the native chromosome is.
- Panel C: The left part of Panel C shows us how SCRaMbLEing a heterodiploid (synIII/III) that does not mate can cause a deletion in part of the synIII chromosome that contained a MATα site. The loss of the MATα site leaves these cells with only one copy of the gene, the MATa allele, and it can therefore now mate. The table on the right part of Panel C describes the frequency of mating types after SCRaMbLEing: for synIII/III strains, 45% became a-maters, while none were α-maters, since it was the MATα site that was lost from synIII in the process. For III/III strains, which were also heterozygous for both MAT alleles, there were no mating cells, since nothing was lost in the process and diploid cells do not mate.