The Method
Before the DNA can be sequenced, it has to be denatured into single strands using heat. Next a primer is annealed to one of the template strands. This primer is specifically constructed so that its 3' end is located next to the DNA sequence of interest. Either this primer or one of the nucleotides should be radioactively or fluorescently labeled so that the final product can be detected on a gel (Russell, 2002). Once the primer is attached to the DNA, the solution is divided into four tubes labeled "G", "A", "T" and "C". Then reagents are added to these samples as follows:
"G" tube: all four dNTP's, ddGTP and DNA polymerase
"A" tube: all four dNTP's, ddATP and DNA polymerase
"T" tube: all four dNTP's, ddTTP and DNA polymerase
"C" tube: all four dNTP's, ddCTP and DNA polymerase
As shown above, all of the tubes contain a different ddNTP present, and each at about one-hundreth the concentration of the the normal precursors (Russell, 2002). As the DNA is synthesized, nucleotides are added on to the growing chain by the DNA polymerase. However, on occasion a dideoxynucleotide is incorporated into the chain in place of a normal nucleotide, which results in a chain-terminating event. For example if we looked at only the "G" tube, we might find a mixture of the following products:

Figure 1: An example of the potential fragments that could be produced in the "G" tube. The fragments are all different lengths due to the random integration of the ddGTP's (Metzenberg).
The key to this method, is that all the reactions start from the same nucleotide and end with a specific base. Thus in a solution where the same chain of DNA is being synthesized over and over again, the new chain will terminate at all positions where the nucleotide has the potential to be added because of the integration of the dideoxynucleotides (Russell, 2002). In this way, bands of all different lengths are produced. Once these reactions are completed, the DNA is once again denatured in preparation for electrophoresis. The contents of each of the four tubes are run in separate lanes on a polyacrylmide gel in order to separate the different sized bands from one another. After the contents have been run across the gel, the gel is then exposed to either UV light or X-Ray, depending on the method used for labeling the DNA.
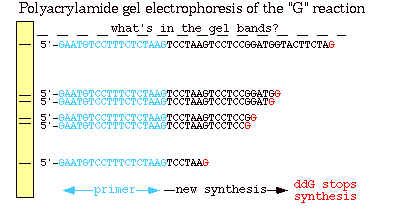
Figure 2: This is a polyacrylmide gel of the reactions in the "G" tube (the same sequences seen in figure 1). The longer fragments of DNA traveled shorter distances than the smaller fragments because of their heavier molecular weight.The blue section indicates the primer, the black section indicates the newly synthesized strand and the red denotes a ddGTP, which terminated the chain (Metzenberg).
As shown in Figure 2, smaller fragments are produced when the ddNTP is added closer to the primer because the chains are smaller and therefore migrate faster across the gel. If all of the reactions from the four tubes are combined on one gel, the actual DNA sequence in the 5' to 3' direction can be determined by reading the banding pattern from the bottom of the gel up. It is important to remember though that this sequence is complementary to the template strand from the beginning.
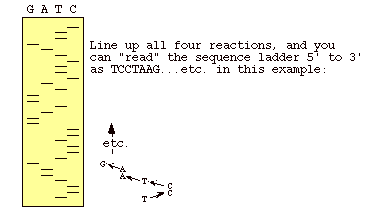
Figure 3: This is an autoradiogram of a dideoxy sequencing gel. The letters over the lanes indicate which dideoxy nucleotide was used in the sample being represented by that lane. When you read from the bottom up, you are reading the complementary sequence of the template strand (Metzenberg).
Automated Sequencing
With the many advancements in technology that we have achieved since 1974, it is no surprise that the Sanger method has become outdated. However, the new technology that has emerged to replace this method is based on the same principles of Sanger's method. Automated sequencing has been developed so that more DNA can be sequenced in a shorter period of time. With the automated procedures the reactions are performed in a single tube containing all four ddNTP's, each labeled with a different color dye (Russell, 2002).
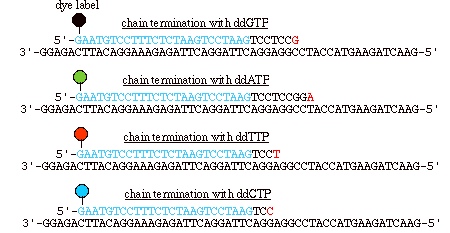
Figure 4: In automated sequencing, the oligonucleotide primers can be "end-labeled" with different color dyes, one for each ddNTP. These dyes fluoresce at different wavelengths, which are read via a machine (Metzenberg).
As in Sanger's method, the DNA is separated on a gel, but they are all run on the same lane as opposed to four different ones.
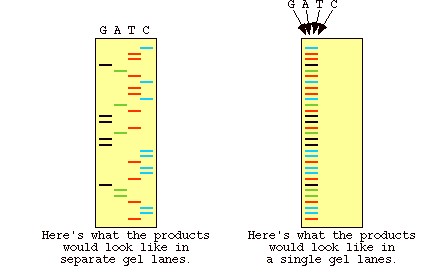
Figure 5: Results of gel electrophoresis for the dye labeled DNA in automated sequencing. The image on the left shows what the gel looks like if the four reactions are run in different lanes, as opposed to the image on the right which shows a gel where all the DNA is run in one lane (Metzenberg).
Since the four dyes fluoresce at different wavelengths, a laser then reads the gel to determine the identity of each band according to the wavelengths at which it fluoresces. The results are then depicted in the form of a chromatogram, which is a diagram of colored peaks that correspond to the nucleotide in that location in the sequence (Russell, 2002).
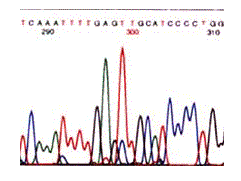
Figure 6: Results from an automated sequence shown in the form of a chromatogram. The colors represent the four bases: blue is C, green is A, black is G and red is T (Metzenberg).
References
1. Russell, Peter. 2002. iGenetics. Pearson Education, Inc., San Francisco, pp 187-189.