
This web page was produced as an assignment for an undergraduate
course at Davidson College.
Reverse Transcriptase Orthologs
Reverse Transcriptase Sequence Conservation in Retroviruses:
Reverse transcriptase (RT) is primarily an RNA directed polymerase that is commonly found in retroviruses, which use RT to make a DNA copy of its own RNA genome. While different retroviruses use different versions of RT's, there are often several conserved similarities of amino acid residues and motifs within the primary structures (Georgiadis et al., 1995). Specifically, there are conserved similarities in RT primary structures of HIV-1, HIV-2, simian immunodeficiency virus (SIV), and Moloney murine leukemia (MoMuLV) retroviruses. The HIV-2 and SIV RT's have the most highly conserved primary structures in comparison with HIV-1 RT, having over 50% of their amino acid residues conserved (See Figures 3 and 5) (NCBI's Blast 2, 2005). Comparison of the MoMuLV RT with HIV-1 RT shows that significant differences exist between the primary structures, but there are still some features that were conserved (See Figures 6 and 7) Specifically, conserved residues lysine 53, glutamine 63, serine 195, and glutamine 260 may have a role in main-chain hydrogen bonding interactions. These residues are often conserved in the RT family and are therefore considered to have significant interactions and structural roles. The overall fold and structures of the finger and palm domains of MoMuLV RT is very similar to those of HIV-1 RT with the exception that the MoMuLV RT sequence includes an additional N-terminal 40 amino acids (Georgiadis et al., 1995). Finally, the trio of aspartic acid residues (110, 185, 186) that are involved in metal ion ligation and phosphate interactions at the polymerase active site of HIV-1 RT (Doublie, Sawaya, & Ellenberger, 1999) are highly conserved in other RT's ( HIV-2, SIV, MoMuLV) (See Figures 3, 4, and 6). Hence, it seems evident that these three aspartic acid residue motifs are necessary in the proper function of the polymerase active site in reverse transcriptases. The RT amino acid sequences shown below were found using Ensembl and Entrez database searches.
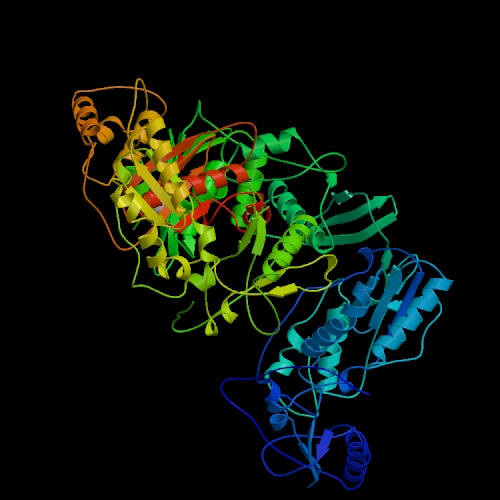
Figure 1. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1 (HIV-1). Image courtesy of Protein Databank. ID: 1HMV; PDB, 2005.
1 PISPIETVPV KLKPGMDGPK VKQWPLTEEK IKALVEICTE MEKEGKISKI
51 GPENPYNTPV FAIKKKDSTK WRKLVDFREL NKRTQDFWEV QLGIPHPAGL
101 KKKKSVTVLD VGDAYFSVPL DEDFRKYTAF TIPSINNETP GIRYQYNVLP
151 QGWKGSPAIF QSSMTKILEP FRKQNPDIVI YQYMDDLYVG SDLEIGQHRT
201 KIEELRQHLL RWGLTTPDKK HQKEPPFLWM GYELHPDKWT VQPIVLPEKD
251 SWTVNDIQKL VGKLNWASQI YPGIKVRQLC KLLRGTKALT EVIPLTEEAE
301 LELAENREIL KEPVHGVYYD PSKDLIAEIQ KQGQGQWTYQ IYQEPFKNLK
351 TGKYARMRGA HTNDVKQLTE AVQKITTESI VIWGKTPKFK LPIQKETWET
401 WWTEYWQATW IPEWEFVNTP PLVKLWYQLE KEPIVGAETF YVDGAANRET
451 KLGKAGYVTN RGRQKVVTLT DTTNQKTELQ AIYLALQDSG LEVNIVTDSQ
501 YALGIIQAQP DQSESELVNQ IIEQLIKKEK VYLAWVPAHK GIGGNEQVDK
551 LVSAGIRKVL
Sequence 1. Complete amino acid sequence of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase.
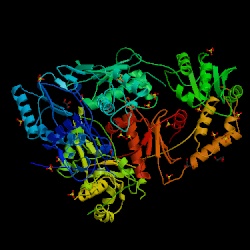
Figure 2. The structure of reverse transcriptase from the human immunodeficiency virus type 2 (HIV-2). Image courtesy of Protein Databank. ID: 1MU2; PDB, 2005.
1 QLEEAPPTNP YNTPTFAIRK KDKNKWRMLI DFRELNKVXX DFTEIQLGIP HPAGLAKKRR
61 ITVLDVGDAY FSIPLHEDFR QYTAFTLPSV NNAEPGKRYI YKVLPQGWKG SPAIFQHTMR
121 QVLEPFRKVN KDVIIIQYMD DILIASDRTD LEHDRVALQL KELLNGLGFS TPDEKFQKDP
181 PYQWMGYELW PTKWKLQKIQ LPQKETWTVN DIQKLVGVLN WAAQLYPGIK TRHLCKLIRG
241 KMTLTEEVQW TDLAEAELEE NKIILSQEQE GRYYQEEKEL EATVQKDQDN QWTYKVHQGE
301 KILKVGKYAK IKNTHTNGVR LLAQ
Sequence 2. Complete amino acid sequence of human immunodeficiency virus type 2 (HIV-2) reverse transcriptase.

Figure 3. Blast 2 results of amino acid sequences from HIV-1 and HIV-2 reverse transcriptases. The boxed regions show conservation of residues at the polymerase active site, particularly the aspartic acids (110, 185, 186). Identities = 187/325 (57%), Positives = 234/325 (71%), Gaps = 1/325 (0%).
1MPRKTGGFFR AWPMGKEAPQ FPHGPDASGA DTNCSXRGSS CGSTEELHEG GQKAEGEQRE
61 TLQGGNGGFA APQFSLWRRP VVTAYIEEQP IEVLLDTGAD DSIVAGIELG PNYTPKIVGG
121 IGGFINTKEY KDVKIKVLGK VIKGTIMTGD TPINIFGRNL LTAMGMSLNL PIAKVEPIKV
181 TLKPGKDGPK LRQWPLSKEK IIALREICEK MEKDGQLEXA PPTNPYNTPT FAIKXXDGNK
241 WRMLIDFREL NKVTQDFTEV QLGIPHPXGL AKRRRITVLD VGDAYFSIPL DEEFRQYTAF
301 TLPSVNNAEP GKRYIYKVLP QGWKGSPAIF QHTXRNVLEP FRKANPDVTL IQYMDDILIA
361 SDRTDLEHDR VVLQLKELLN SIGFSTPEEK FQKDPPFQWM GYELWPTKWK LQKIELPQRE
421 TWTVNNIQKL VGVLNWAAQI YPGIKTKHLC RLIRGKMTLT EEVQWTEMAE AEYEENKIIP
481 SQEPEGCYYQ EGKPLEAPVI KSQDNQWSYK IHQEDKILKV GKFAKIKNTH TNGVSLLAHV
541 VQKIGKEAII IWGQVPRFHL PVEXEIWEQW WTDYWQVTWI PEWDFVSTPP LVRLVFNLVK
601 EPIQGAETFY VDRSCNRQSR EGKAGYVTDR GRGKTKLLEQ TTNQQAELEA FYLALADSGP
661 KANIIVDSQY VMGIVAGQPT ESESRLVNQI IEKMIKKEAI YVAWVPAHKX IGENQEVDHL
721 VSQEIRQVLF LEKIEPAQEE HEKYHSNVKK LVFKFXLPRL VAKQIVDTCD KCHLKGEAIH
781 GQVNAXLGTW QMDCTHLEGK IIIVAVHVGS GFIEAEVIPX ETGRQTALFL LKLASRWPIT
841 HLHTDNGANF TSQEVKMVAW WAGIEQTFXV PYNPQSQXVV EAMNHHLKTQ IDRIREQANS
901 IETIVLMAIH CINFKRRGGI GDMTPAERLV NMITTEQEIQ FQQSKNSKFK NFRVYYREGR
961 DQLWKGPGEL LWKGEGAVIL KVGTEIKVVP RRKAKIIKDY GGGKELDSGS HLEDTGEARE
1021 VA
Sequence 3. Complete amino acid sequence of simian immunodeficiency virus (SIV) reverse transcriptase.

Figure 4. Blast 2 results of amino acid sequences from HIV-1 and SIV reverse transcriptases. The boxed regions show conservation of residues at the polymerase active site, particularly the aspartic acids (110, 185, 186). Identities = 324/560 (57%), Positives = 409/560 (72%), Gaps = 1/560 (0%).
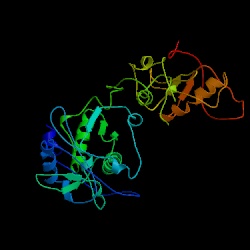
Figure 5. The structure of reverse transcriptase from the human immunodeficiency virus type 2. Image courtesy of Protein Databank. ID: 1RW3; PDB, 2005.
1 GGQGQEPPPE PRITLKVGGQ PVTFLVDTGA QHSVLTQNPG PLSDKSAWVQ GATGGKRYRW
61 TTDRKVHLAT GKVTHSFLHV PDCPYPLLGR DLLTKLKAQI HFEGSGAQVM GPMGQPLQVL
121 TLNIEDEHRL HETSKEPDVS LGSTWLSDFP QAWAETGGMG LAVRQAPLII PLKATSTPVS
181 IKQYPMSQEA RLGIKPHIQR LLDQGILVPC QSPWNTPLLP VKKPGTNDYR PVQDLREVNK
241 RVEDIHPTVP NPYNLLSGLP PSHQWYTVLD LKDAFFCLRL HPTSQPLFAF EWRDPEMGIS
301 GQLTWTRLPQ GFKNSPTLFD EALHRDLADF RIQHPDLILL QYVDDLLLAA TSELDCQQGT
361 RALLQTLGNL GYRASAKKAQ ICQKQVKYLG YLLKEGQRWL TEARKETVMG QPTPKTPRQL
421 REFLGTAGFC RLWIPGFAEM AAPLYPLTKT GTLFNWGPDQ QKAYQEIKQA LLTAPALGLP
481 DLTKPFELFV DEKQGYAKGV LTQKLGPWRR PVAYLSKKLD PVAAGWPPCL RMVAAIAVLT
541 KDAGKLTMGQ PLVILAPHAV EALVKQPPDR WLSNARMTHY QALLLDTDRV QFGPVVALNP
601 ATLLPLPEEG LQHNCLDILA EAHGTRPDLT DQPLPDADHT WYTDGSSLLQ EGQRKAGAAV
661 TTETEVIWAK ALPAGTSAQR AELIALTQAL KMAEGKKLNV YTDSRYAFAT AHIHGEIYRR
721 RGLLTSEGKE IKNKDEILAL LKALFLPKRL SIIHCPGHQK GHSAEARGNR MADQAARKAA
781 ITETPDTSTL LIENSSPYTS EHFHYTVTDI KDLTKLGAIY DKTKKYWVYQ GKPVMPDQFT
841 FELLDFLHQL THLSFSKMKA LLERSHSPYY MLNRDRTLKN ITETCKACAQ VNASKSAVKQ
901 GTRVRGHRPG THWEIDFTEI KPGLYGYKYL LVFIDTFSGW IEAFPTKKET AKVVTKKLLE
961 EIFPRFGMPQ VLGTDNGPAF VSKVSQTVAD LLGIDWKLHC AYRPQSSGQV ERMNRTIKET
1021 LTKLTLATGS RDWVLLLPLA LYRARNTPGP HGLTPYEILY GAPPPLVNFP DPDMTRVTNS
1081 PSLQAHLQAL YLVQHEVWRP LAAAYQEQLD RPVVPHPYRV GDTVWVRRHQ TKNLEPRWKG
1141 PYTVLLTTPT ALKVDGIAAW IHAAHVKAAD PGGGPSSRLT WRVQRSQNPL KIRLTREAP
Sequence 4. Complete amino acid sequence of Moloney murine leukemia virus (MoMuLV) reverse transcriptase.

Figure 6. Blast 2 results of amino acid sequences from HIV-1 and Moloney MuLV reverse transcriptases. The boxed regions show conservation of residues at the polymerase active site, particularly the aspartic acids (110, 185, 186). Identities = 73/275 (26%), Positives = 130/275 (46%), Gaps = 19/275 (6%).
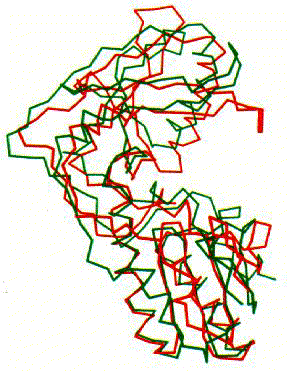
Figure 7. Comparison of MoMuLV RT and HIV-1 RT fingers and palm domains using superimpositions. Traces are shown for MoMuLV RT (green) and HIV-1 RT (red). Image courtesy of Structure.
Reverse Transcriptase Structural Similarities with DNA Polymerases:
Reverse transcriptases are usually much more structurally simple than other more conventional replicative polymerases, but are still very effective in performing their function. In addition, their absence of editing machinery allows mutations that are advantageous in escaping immune defenses (Georgiadis et al., 1995). The amino acid sequence of HIV-1 RT was compared with DNA polymerases from human, mouse, rat, E. coli, yeast, Thermus Aquaticus, and T7 bacteriophage using NCBI's Blast 2, and no significant conserved similarities were found (NCBI's Blast 2, 2005). Although polymerases often do not share any significant conserved similarities in primary structure (with the exception of humans, mice, and rats), they are often found to be structurally similar, usually having a DNA binding mechanism comprising "finger," "palm," and "thumb" subdomains (See Figures 8 and 9) (Doublie, Sawaya, & Ellenberger, 1999). Furthermore, although the primary structures of these polymerases are not conserved, they all are able to fulfill their primary function of polymerizing DNA strands. The amino acid sequences and illustrated figures of several different DNA polymerases are shown below to demonstrate the wide range and dissimilarity of primary structures, yet some similarities shared in overall structures. Sequences were found using Ensembl and Entrez database searches.
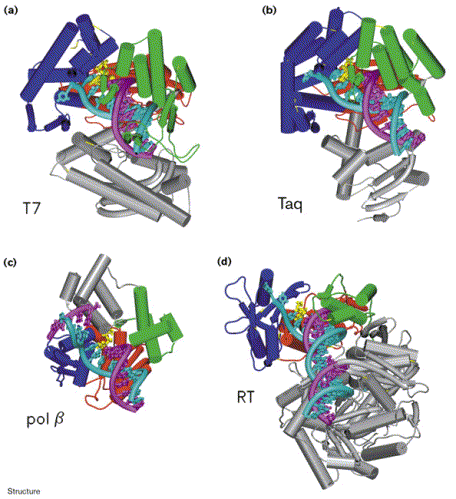
Figure 8. Comparison of the structures of four different DNA polymerases in complex with DNA: (a) T7 DNA polymerase, (b) Taq polymerase, (c) pol b, and (d) HIV-1 Reverse Transcriptase (p66 subunit only). DNA is shown in cyan and magenta, and the nucleotide substrates in yellow. The fingers subdomain is shown in dark blue, the thumb in green, and the palm in red. Image courtesy of Structure.
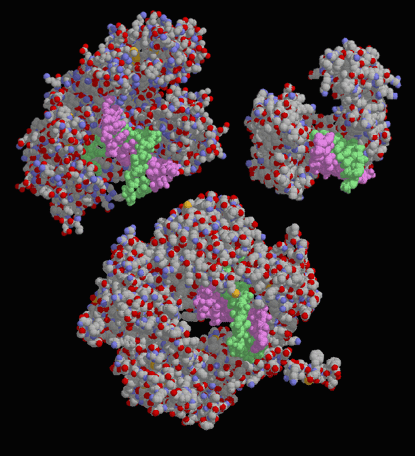
Figure 9. Drawings of three different DNA polymerases in complex with a DNA strand. At upper left is DNA polymerase I from Escherichia coli, upper right is human DNA polymerase, and at bottom is a viral DNA polymerase. Notice that the three structures are quite different in shape and size, but all are able to specifically bind and polymerize DNA. Image courtesy of David S. Goodsell.
. 
Figure 10. The structure of DNA polymerase in humans (Homo sapien). Image courtesy of Protein Databank. ID: 1TV9; PDB, 2005. Notice that mice (Mus musculus) and rats (Rattus norvegicus) have the same exact primary sequence, and hence share the same 3-dimensional structure as well.
1 MSKRKAPQETLNGGITDMLTELANFEKNVSQAIHKYNAYRKAASVIAKYPHKIKSGAEAK
61 KLPGVGTKIAEKIDEFLATGKLRKLEKIRQDDTSSSINFLTRVSGIGPSAARKFVDEGIK
121 TLEDLRKNEDKLNHHQRIGLKYFGDFEKRIPREEMLQMQDIVLNEVKKVDSEYIATVCGS
181 FRRGAESSGDMDVLLTHPSFTSESTKQPKLLHQVVEQLQKVHFITDTLSKGETKFMGVCQ
241 LPSKNDEKEYPHRRIDIRLIPKDQYYCGVLYFTGSDIFNKNMRAHALEKGFTINEYTIRP
301 LGVTGVAGEPLPVDSEKDIFDYIQWKYREPKDRSE
Sequence 5. Complete amino acid sequence of DNA polymerase in humans (Homo sapiens).
1 MSKRKAPQETLNGGITDMLVELANFEKNVSQAIHKYNAYRKAASVIAKYPHKIKSGAEAK
61 KLPGVGTKIAEKIDEFLATGKLRKLEKIRQDDTSSSINFLTRVTGIGPSAARKFVDEGIK
121 TLEDLRKNEDKLNHHQRIGLKYFEDFEKRIPREEMLQMQDIVLNEIKKVDSEYIATVCGS
181 FRRGAESSGDMDVLLTHPNFTSESSKQPKLLHRVVEQLQKVHFITDTLSKGETKFMGVCQ
241 LPSEKDGKEYPHRRIDIRLIPKDQYYCGVLYFTGSDIFNKNMRAHALEKGFTINEYTIRP
301 LGVTGVAGEPLPVDSEQDIFDYIQWRYREPKDRSE
Sequence 6. Complete amino acid sequence of DNA polymerase in mice (Mus musculus).
1 MSKRKAPQETLNGGITDMLVELANFEKNVSQAIHKYNAYRKAASVIAKYPHKIKSGAEAK
61 KLPGVGTKIAEKIDEFLATGKLRKLEKIRQDDTSSSINFLTRVTGIGPSAARKLVDEGIK
121 TLEDLRKNEDKLNHHQRIGLKYFEDFEKRIPREEMLQMQDIVLNEVKKLDPEYIATVCGS
181 FRRGAESSGDMDVLLTHPNFTSESSKQPKLLHRVVEQLQKVRFITDTLSKGETKFMGVCQ
241 LPSENDENEYPHRRIDIRLIPKDQYYCGVLYFTGSDIFNKNMRAHALEKGFTINEYTIRP
301 LGVTGVAGEPLPVDSEQDIFDYIQWRYREPKDRSE
Sequence 7. Complete amino acid sequence of DNA polymerase in rats (Rattus norvegicus).
Figure 11. The structure of DNA polymerase in bacteria (Escherichia coli ). Image courtesy of Protein Databank. ID: 1Q8I; PDB, 2005.
1 MAQAGFILTR HWRDTPQGTE VSFWLATDNG PLQVTLAPQE SVAFIPADQV PRAQHILRGE
61 QGFRLTPLAL KDFHRQPVYG LYCRAHRQLM NYEKRLREGG VTVYEADVRP PERYLMERFI
121 TSPVWVEGDM HNGAIVNARL KPHPDYRPPL KWVSIDIETT RHGELYCIGL EGCGQRIVYM
181 LGPENGDASA LDFELEYVAS RPQLLEKLNA WFANYDPDVI IGWNVVQFDL RMLQKHAERY
241 RIPLRLGRDN SELEWREHGF KNGVFFAQAK GRLIIDGIEA LKSAFWNFSS FSLETVAQEL
301 LGEGKSIDNP WDRMDEIDRR FAEDKPALAT YNLKDCELVT QIFHKTEIMP FLLERATVNG
361 LPVDRHGGSV AAFGHLYFPR MHRAGYVAPN LGEVPPHASP GGYVMDSRPG LYDSVLVLDY
421 KSLYPSIIRT FLIDPVGLVE GMAQPDPEHS TEGFLDAWFS REKHCLPEIV TNIWHGRDEA
481 KRQGNKPLSQ ALKIIMNAFY GVLGTTACRF FDPRLVSSIT MRGHQIMRQT KALIEAQGYD
541 VIYGDTDSTF VWLKGAHSEE EATKIGRALV QHVNVWWAET LQKQQLTSAL ELEYETHFCR
601 FLMPTIRGAD TGSKKRYAGL IQEGDKQRMV FKGLETVRTD WTPLAQQFQQ ELYLRIFRNE
661 PYQEYFRETI DKLMAGELDA RLVYRKRLRR PLSEYQRNVP PHVRAARLAD EENQKRGRPL
721 QYQNRGTIKY VWTTNGPEPL DYQRSPLDYE HYLTRQLQPV AEGILPFIED NFATLMTGQL
781 GLF
Sequence 8. Complete amino acid sequence of DNA polymerase in prokaryotic bacteria (Escherichia coli ).
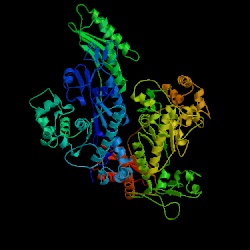
Figure 12. The structure of DNA polymerase in yeast (Saccharomyces cerevisiae). Image courtesy of Protein Databank. ID: 1JIH; PDB, 2005.
1 MSLKGKFFAF LPNPNTSSNK FFKSILEKKG ATIVSSIQNC LQSSRKEVVI LIEDSFVDSD
61 MHLTQKDIFQ REAGLNDVDE FLGKIEQSGI QCVKTSCITK WVQNDKFAFQ KDDLIKFQPS
121 IIVISDNADD GQSSTDKESE ISTDVESERN DDSNNKDMIQ ASKPLKRLLQ GDKGRASLVT
181 DKTKYKNNEL IIGALKRLTK KYEIEGEKFR ARSYRLAKQS MENCDFNVRS GEEAHTKLRN
241 IGPSIAKKIQ VILDTGVLPG LNDSVGLEDK LKYFKNCYGI GSEIAKRWNL LNFESFCVAA
301 KKDPEEFVSD WTILFGWSYY DDWLCKMSRN ECFTHLKKVQ KALRGIDPEC QVELQGSYNR
361 GYSKCGDIDL LFFKPFCNDT TELAKIMETL CIKLYKDGYI HCFLQLTPNL EKLFLKRIVE
421 RFRTAKIVGY GERKRWYSSE IIKKFFMGVK LSPRELEELK EMKNDEGTLL IEEEEEEETK
481 LKPIDQYMSL NAKDGNYCRR LDFFCCKWDE LGAGRIHYTG SKEYNRWIRI LAAQKGFKLT
541 QHGLFRNNIL LESFNERRIF ELLNLKYAEP EHRNIEWEKK TA
Sequence 9. Complete amino acid sequence of DNA polymerase in yeast (Saccharomyces cerevisiae).
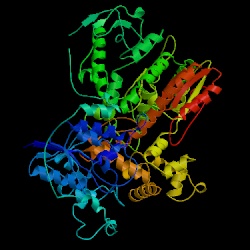
Figure 13. The structure of Taq polymerase (Thermus aquaticus). Image courtesy of Protein Databank. ID: 1CMW; PDB, 2005.
1 MRGMLPLFEP KGRVLLVDGH HLAYRTFHAL KGLTTSRGEP VQAVYGFAKS LLKALKEDGD
61 AVIVVFDAKA PSFRHEAYGG YKAGRAPTPE DFPRQLALIK ELVDLLGLAR LEVPGYEADD
121 VLASLAKKAE KEGYEVRILT ADKDLYQLLS DRIHVLHPEG YLITPAWLWE KYGLRPDQWA
181 DYRALTGDES DNLPGVKGIG EKTARKLLEE WGSLEALLKN LDRLKPAIRE KILAHMDDLK
241 LSWDLAKVRT DLPLEVDFAK RREPDRERLR AFLERLEFGS LLHEFGLLES PKALEEAPWP
301 PPEGAFVGFV LSRKEPMWAD LLALAAARGG RVHRAPEPYK ALRDLKEARG LLAKDLSVLA
361 LREGLGLPPG DDPMLLAYLL DPSNTTPEGV ARRYGGEWTE EAGERAALSE RLFANLWGRL
421 EGEERLLWLY REVERPLSAV LAHMEATGVR LDVAYLRALS LEVAEEIARL EAEVFRLAGH
481 PFNLNSRDQL ERVLFDELGL PAIGKTEKTG KRSTSAAVLE ALREAHPIVE KILQYRELTK
541 LKSTYIDPLP DLIHPRTGRL HTRFNQTATA TGRLSSSDPN LQNIPVRTPL GQRIRRAFIA
601 EEGWLLVALD YSQIELRVLA HLSGDENLIR VFQEGRDIHT ETASWMFGVP REAVDPLMRR
661 AAKTINFGVL YGMSAHRLSQ ELAIPYEEAQ AFIERYFQSF PKVRAWIEKT LEEGRRRGYV
721 ETLFGRRRYV PDLEARVKSV REAAERMAFN MPVQGTAADL MKLAMVKLFP RLEEMGARML
781 LQVHDELVLE APKERAEAVA RLAKEVMEGV YPLAVPLEVE VGIGEDWLSA KE
Sequence 10. Complete amino acid sequence of DNA polymerase (Taq polymerase) in Thermus aquaticus
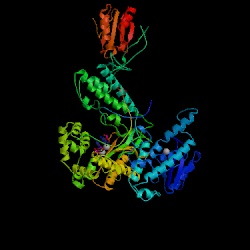
Figure 14. The structure of DNA polymerase in T7 Bacteriophage. Image courtesy of Protein Databank. ID: 1SKR; PDB, 2005.
1 MIVSDIEANA LLESVTKFHC GVIYDYSTAE YVSYRPSDFG AYLDALEAEV ARGGLIVFHN
61 GHKYDVPALT KLAKLQLNRE FHLPRENCID TLVLSRLIHS NLKDTDMGLL RSGKLPGKRF
121 GSHALEAWGY RLGEMKGEYK DDFKRMLEEQ GEEYVDGMEW WNFNEEMMDY NVQDVVVTKA
181 LLEKLLSDKH YFPPEIDFTD VGYTTFWSES LEAVDIEHRA AWLLAKQERN GFPFDTKAIE
241 ELYVELAARR SELLRKLTET FGSWYQPKGG TEMFCHPRTG KPLPKYPRIK TPKVGGIFKK
301 PKNKAQREGR EPCELDTREY VAGAPYTPVE HVVFNPSSRD HIQKKLQEAG WVPTKYTDKG
361 APVVDDEVLE GVRVDDPEKQ AAIDLIKEYL MIQKRIGQSA EGDKAWLRYV AEDGKIHGSV
421 NPNGAVTGRA THAFPNLAQI PGVRSPYGEQ CRAAFGAEHH LDGITGKPWV QAGIDASGLE
481 LRCLAHFMAR FDNGEYAHEI LNGDIHTKNQ IAAELPTRDN AKTFIYGFLY GAGDEKIGQI
541 VGAGKERGKE LKKKFLENTP AIAALRESIQ QTLVESSQWV AGEQQVKWKR RWIKGLDGRK
601 VHVRSPHAAL NTLLQSAGAL ICKLWIIKTE EMLVEKGLKH GWDGDFAYMA WVHDEIQVGC
661 RTEEIAQVVI ETAQEAMRWV GDHWNFRCLL DTEGKMGPNW AICH
Sequence 11. Complete amino acid sequence of DNA polymerase in T7 Bacteriophage.
Conclusion:
Although reverse transcriptases are considered to be types of DNA polymerases, it is clear that the amino acid sequences of RT's are not conserved among other more conventional replicative polymerases. There is, however, significant conservation of sequences among different types of reverse transcriptases, particularly those that have a key role in specific structure and function. Still, no matter how dissimilar in conservation of sequences, polymerases (RT's included) are able to perform their function of polymerizing DNA strands. It seems evident that although primary structure can often hold the key to a protein's function, there may be several different ways for an enzyme to be built and still carry out its specific purpose.
Doublie S, Sawaya MR, Ellenberger T. 1999. An open and closed case for all
polymerases. Structure (7)2. <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=B-WA-A-W-AB-MsSAYWA-UUA-AAAAUVBBVD-AAUYZWVAVD-EWAVDBCAC-AB-U&_rdoc=4&_fmt=full&_udi=B6VSR-4CXCWVD-T&_coverDate=02%2F15%2F1999&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&_
version=1&_urlVersion=0&_userid=2665120&md5=260675e1d2914c88e9789eb57c0e2e4f>
Accessed 2005 Feb 12.
Georgiadis MM, Jessen SM, Ogata CM, Telesnitsky A, Goff SP, Hendrickson WA.
1995. Mechanistic implications from the structure of a catalytic fragment of
Moloney murine leukemia virus reverse transcriptase. Structure (3)9. <http://www.sciencedirect.com/science?_ob=ArticleURL&_aset=V-WA-A-W-AB-MsSAYWA-UUW-U-AAAWEWWYBB-AAAUCUBZBB-EDUADEEUU-AB-U&_rdoc=9&_fmt=full&_udi=B6VSR-4CXD8FR-3H&_coverDate=09%2F30%2F1995&_cdi=6269&_orig=search&_st=13&_sort=d&view=c&_acct=C000058476&_
version=1&_urlVersion=0&_userid=2665120&md5=9873b2e5d34167f8a489d9dc556cffdc>
Accessed 2005 March 4.
NCBI's BLAST2. National Center for Biotechnology Information. <http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html> Accessed 2005 March 4.
Davidson College Molecular Biology Homepage
My Molecular Biology Home Page
© Copyright 2005 Department of Biology, Davidson College,
Davidson, NC 28035
Send comments, questions, and suggestions to: kekoike@davidson.edu;
macampbell@davidson.edu