*This website was produced as an assignment for an undergratuate course at
Davidson College.*
Orthologs of Alcohol Dehydrogenase
Orthologs
Orthologs are genes in different species that have evolved from a common ancestor. Usually, orthologs conserve similar gene sequences and functions in the different organisms over time. Determining orthologs allows us to more precisely discover similarities and differences among closely related species; two species that are closely related should have very similar orthologs. Furthermore, orthologs may allow us to predict the function of newly sequenced genomes.
Types of Alcohol Dehydrogenase
Jornvall et al (1987) have posited a manner in which the various alcohol dehydrogenases have come about. The early events involved duplications and recombinations that led to long dehydrogenases with zinc and short dehydrogenases without zinc (See previous website for ADH structure). Later duplications led to a separation of alcohol dehydrogenase and sorbital dehydrogenases. These duplications were followed by further duplications (specifically in the ADH group) to produce separate classes and separate subunits in those classes (Figure 1).
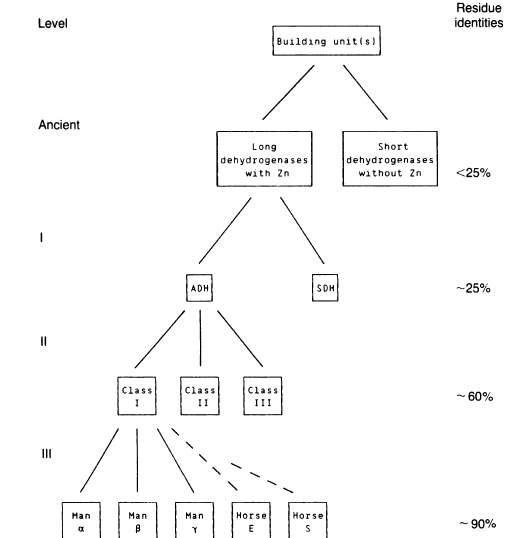
Figure 1. A flow scheme to demonstrate the relationships of the different alcohol dehydrogenases. Permission pending Jornvall et al.
More specifically, Jornvall et al. classified five groups of alcohol dehydrogenases along with their sub-classes (if applicable):
1. Mammalian/plant ADHs
a. Class I: Horse liver alcohol dehydrogenase and the three human ADH subunit types α, β, γ
b. Class II: Two isoenzymes from maize and human ADH subunit Φ
c. Class III: Avian ADH and the human ADH subunit χ
d. Class IV: Human ADH6
2. Bacterial ADHs
3. Tetrameric yeast ADHs
4. Polyol dehydrogenases
5. E. coli threonine dehydrogenase
Conservation Among Isoenzymes
Homo Sapiens
Below are the ClustalW2 alignment results of three classes of human ADH (Figure 2). While there is amino acid conservation among all three shown classes of human ADH, the β and γ subunits of Class I show the most conservation. While the amino acid sequence of the α subunit could not be found, it should be noted that it had 95.1% homology with the β subunit and 94.0% homology with γ subunit. On the other hand, the Class II ADH only had a 60-65% homology with the three subunits of Class I ADHs. These differences in gene sequence may be explained by the different locations of the ADHs. Class II ADH is found mainly in the liver and stomach while the Class III ADH is expressed in many tissues and is the only ADH in the brain (Cotton & Goldman, 1988).

Figure 2. ClustalW2 alignment results of human ADH Class I (Β and γ subunits), Class II, and Class III.
Saccharomyces cerevisiae
Below is the ClustalW2 alignment for three ADH isoenyzmes of S. cerevisiae (Figure 3). As can be seen, there is most conservation among the SADHI and SADH II isoenzymes. They differ by only 23 amino acids, but this change explains the two isoenzymes' difference for substrate affinity: SADHII has a lower substrate affinity for ethanol than does SADHI. These two isoenzymes also carry out different metabolic roles. SADHI is the fermentative enzyme that can account for up to 1% of cell protein, while SADHII converts ethanol to acetaldehyde. SADHIII is located in the mitochondria of the yeast and has 79 and 78% identical amino acid sequence with SADHI and SADHII respectively. However, its functnion has not been identified (Reid & Fewson 1994).
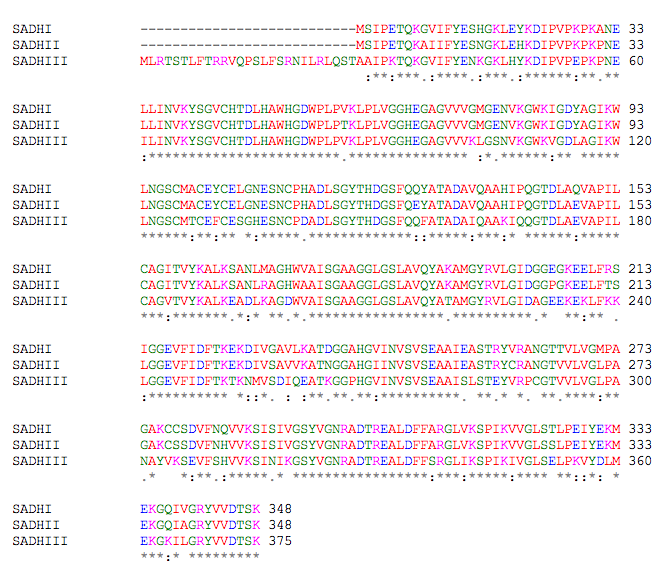
Figure 3. ClustalW2 alignment for three isoenzymes of the yeast S. cerevisiae.
Conservation Among Different Species
A ClustalW2 alignment of the amino acid sequences of organisms in the Big 7 with ADH is shown below (Figure 4). Although ADH is known to exist in some plants, ADH in Arabidopsis was searched for in the ClustalW2 database and in multiple review papers, but none could be found. An ADH-related enzyme was found for Escherichia coli and will be discussed in Table I.
By looking at Figure 4, one can see that there are many identical amino acids between mouse and human ADH sequences. It has previously been identified that the human ADHI structure has been conserved in the rat and mouse ADH genes (Hittle & Crabb, 1988). The mouse has three separate classes of ADH isoenzymes on chromosome 3 which have similar properties like the three classes of human ADH isoenzymes (Cotton & Goldman, 1988). Conversely, there are barely any identical amino acids in Drosophila compared to the all of the other orthologs. Its ADH enzyme is classified as a short dehydrogenase without zinc; its alignment shows how much fewer amino acids it has compared to the other orthologs, and as a result of its very few amino acids, it does not contain any zinc binding sites. In addition to alcohols, these enzymes can also use steroids, glucose, and precurosors for antibiotics as substrates (Heinstra, 1993).
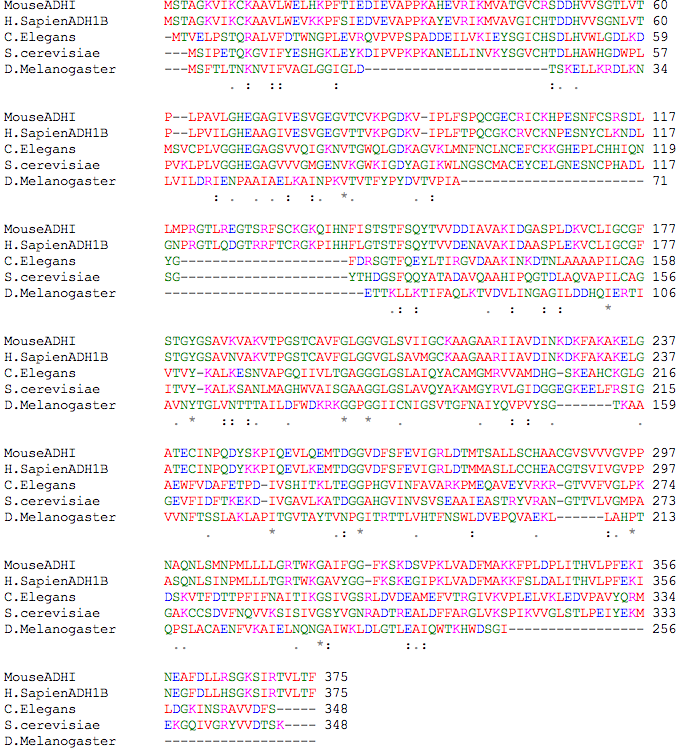
Figure 4. ClustalW2 Alignment for organisms of the Big 7 with identified ADH enzymes.
As previously mentioned, E. coli does not contain an alcohol dehydrogenase, but it does have an ADH-related enzyeme known as Threonine dehydrogenase that forms a tetrameric structure and is NAD(H) dependent (like all of the other previously mentioned orthologs). Table 1 shows the similarities in amino acid sequences for multiple ADH enzymes in different organisms. The ADH-related enzyme forE. coli only has 27 similar amino acids with horse liver alcohol dehydrogenase (which is very similar to human ADH), 30 amino acids with SADHI, 29 amino acids with SADHII, and 30 amino acids with SADHIII.
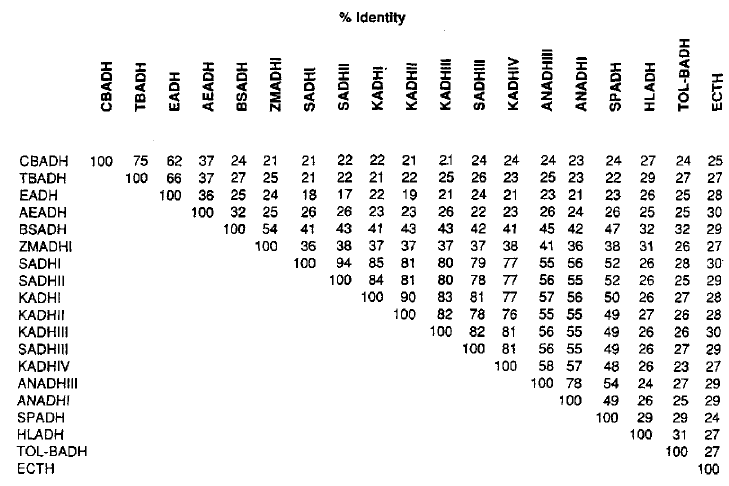
Table I. Comparison of identical amino acids in a selection of ADH enzymes. Permission pending Reid & Fewson (1994).
Despite these varying differences, all NAD(H) dependent enzymes have shown to have the Asp-223 amino acid, which is identified as being responsible for the coenzyme binding of NAD(H) in the active site as well as responsible for prohibiting NADP(H) from binding. Thus, it appears this is conserved because it is vital for coenzyme specificity (Reid & Fewson, 1988).
References
Cotton, R, Goldman, D. 1988. Reviw of the molecular biology of the human alcohol dehydrogenase genes and gene products. Advance in Alcohol & Substance Abuse; 7 (3-4): 171-182. http://www.ncbi.nlm.nih.gov/pubmed/3066190
Heinstra, P. 1993. Evolutionary genetics of the Drosophila alcohol dehydrogenase gene-enzyme system. Genetica; 92: 1-22. http://www.ncbi.nlm.nih.gov/sites/entrez
Jornvall, H, Hoog,J, von Bahr-Lindstrom, H, Vallee, B. 1987 May. Mammalian alcohol dehydrogenases of separate classes: Intermediates between different enzymes and intraclass isozymes. Biochemistry; 84: 2580-2584. http://www.ncbi.nlm.nih.gov/sites/entrez
Reid, M, Fewson, C. 1994. Molecular characterization of microbial alcohol dehydrogenases. Critical Reviews in Microbiology; 20 (1): 13-56. http://www.ncbi.nlm.nih.gov/pubmed/8185833
Ensembl. http://uswest.ensembl.org/index.html
ClustalW2. http://www.ebi.ac.uk/Tools/clustalw2/index.html
Return to my molecular page
Molecular Biology Main Page
Biology Main Page
© Copyright 2010 Department of Biology, Davidson College,
Davidson, NC 28035
Send comments, questions, and suggestions to: Marissa Ghant at maghant@davidson.edu




