*This website was produced as an assignment for an undergratuate course at Davidson College.*
My Favorite Insulin Orthologs

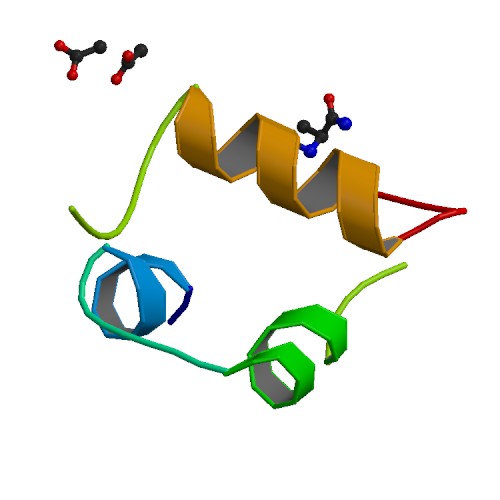
Figure 1. Structure of insuline courtesy of Protein Data Bank
Background
Orthologs are homologous proteins for different species that usually arise from a single evolutionary constant. Insulin is an important study for orthologs because of diabetes. To treat diabetes, insulin is injected into the patient, but it is difficult to make and excess of human insulin. However, there are many orthologs of insulin similar enough to human insulin that when injected, there is no immune response.
Things to Keep in Mind About Insulin
Insulin has two peptide chains containing three disulfide bridges. There are also five areas of insulin that are known to interact with receptors Figure 2 (Greaves et al., 2004). In order for orthologs to be viable replacements for human insulin, the disulfide bridges and binding areas should be in tact.
![]()
Figure 2. Amino acid sequence (in single letter code) of insulin with binding site undelined (Greaves et al., 2004).
Orthologs of Insulin
Insulin like peptides in invertebrates: Invertebrates do not produce mammalian insulin. However, they do produce insulin like peptides that serve in similar pathways as insulin. For many invertebrates, there are multiple insulin like proteins. Drosophila melanogaster has 7 insulin like proteins which suggests that there are different pathways for each insulin like protein (Gronke et al., 2010).
Insulin in fish: Insulin in fish, specifically cod, is quite similar to mammilian insulin. Both proteins contain 51 amino acids and three disulfide bridges. However, the amino acids in the peptide chains are not exactly the same. It seems that cod insulin contains amino acids like methionine and proline in excess compared to that of bovine insulin. Furthermore, cod insulin has aspartic acids as its main acidic amino acid whereas bovine insulin has glutamic acid as its main acidic amino acid (Grant et al. 1967).
Insulin in mammals: The most similar insulin ortholog to human insulin is porcine insulin. Porcine insulin only differs from human insulin by one amino acid at the C-terminus of the B chain (Figure 3). The fact that porcine insulin and human insulin only differ by one amino acid suggests that insulin is conserved through evolution. However much larger differences in mammalian insulin orthologs exist. For example caviomorph rodents produce two different insulin proteins thought to be conserved from either the Cavioidea and the Chinchilloidea families or the Octodontoidea familie (Opazo et al., 2005). The insulin conserved from the Cavioidea and the Chinchilloidea families maintians the 51 amino acid structure with minor differences in specific amino acids. The insulin originating from the Octodontoidea family has a phenylalinin deletion as well as two amino acid insertions on the A chain at the C-terminus (Figure 4) (Opazo et al., 2005). There are also mammalian insulin-like peptides that maintain certain attributes necessary for insulin to be effective in its pathways. One example of such a peptide is the Leydig insulin-like peptide from boar testis. These peptides conserve a couple important factors that all proteins in the insulin superfamily have including a signal peptide, A and B chains and connecting disulfide bridges (Adham et al., 1993). Figure 5 shows some similarities between the Leydig insulin-like and other proteins in the insulin superfamily.

Figure3. This is a comparison between human porcine (pig) and cow insulin. Note that there is only one difference between human and porcine insulin and only three amino acid differenced for cows. Also keep in mind that the differences do not affect the binding site od disulfide bridges. Image from http://www.etetens.de/biobili/images/insuling2.gif permission pending.
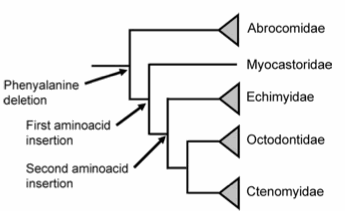
Figure 4. Evolutionay tree based on the phenylalanine deletion as well as two aminoacid insertions. Notice that only the Octodontidae family and not the Cavioidea and the Chinchilloidea families are referenced bacause the Cavioidea and the Chinchilloidea familiesare not missing the phenylalanine (Opazo et al., 2005).
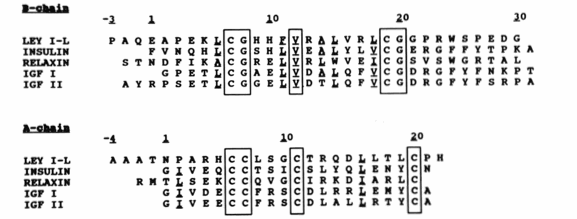
Figure 5. Comparison between Leydig insulin-like peptide and other proteins from the insulin superfamily. Notice that they all have the cysteine peptide for disulfide bridges. The underlined valine peptide causes the center of the protein to be hydrophbic (Adham et al., 1993).
Conclusions
Insulin orthologs are produced in many different animals. Be it invertibrates, fish or mammals, they all at least have insulin-like proteins that are made of two chains connected by disulfide bridges and a hydrophobic center.It is not known which organism predated all the rest and gave rise to insulin, but we can assume that insulin has been conserved fairly well through evolution because there are organisms that maintian the most minute changes that allow us to follow insulin's evolution.
References
Adham I., Burkhardt E., Benahmed M., Engel W. Cloning of a cDNA for a Novel Insulin-like Peptide of the Testicular Leydig Cells. The Journal of Biological Chemistry 268, 26668-26672. 1993.(PDF)
Grant P., Reid K. Isolation and a Partial Amino Acid Sequence of Insulin from the Islet Tissue of Cod (Gadus callarias). Biochem 106, 531-541. 1967. (PDF)
Greaves R., Dosdon G., Verma C. Altered ionization of the B13 Glu in insulin B9 and B10 mutants: a computational analysis. Protein Engineering, Design & Selection. 17, 557-563. 2004.(PDF)
Gronke S., Clarke D., Broughton S., Andrews T., Partridge L. Molecular Evolution and Functional Characterization of Drosophila Insulin-Like Peptides. Plos genetics 6, 1-18. 2010.
Opazo J., Palma R., Melo F., Lessa E. Adaptive Evolution of the Insulin Gene in Caviomorph Rodents. Oxford Journals, 1290-1298. 2005. (PDF)
http://www.etetens.de/biobili/images/insuling2.gif accessed march 7 2010.
Questions? Contact me at Joholzwarth@davidson.edu
Last updated March 10, 2010
Please direct questions or comments to my email name at davidson dot edu