This web page was produced as an assignment for an undergraduate course at Davidson College.
SGS1 and YMR187C: Longevity in Yeast?
Overview:
SGS1/YMR190C, also known as TPS1, is a gene found on Saccharomyces cerevisiae (yeast) chromosome XIII (see Fig. 1 below). SGS1 is a nucleolar DNA helicase of the RecQ family. RecQ is a helicase found in Escherichia coli, and the RecQ family helicases are important for maintaining genome integrity and coordinating genome maintenance pathways in living cells (Bennett and Keck 2004). To see a graphical relationship of the RecQ family, click here. To view the clusters of orthologous groups for the RecQ proteins, click here. The SGS1 helicase is similar to the human BLM and WRN helicases, which, when mutated, can result in cancer-prone Bloom's Syndrome and premature-aging Werner's Syndrome, respectively (SGD 2004).
YMR187C is a hypothetical open reading frame (ORF) located just upstream of SGS1. Its molecular and biological functions have not yet been characterized.

Figure 1: Map of Saccharomyces cerevisiae chromosome XIII features that span coordinates 630914 - 655257 bp. The SGS1 open reading frame stretches from 645257 to 640914 bp on the Crick strand (shown in the 3' to 5' direction). The nearest non-annotated gene, YMR187C, is from 635983 to 634688 bp, also on the Crick strand. See the following link to SGD 2004 for map key, notes, and more information: http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004802.
SGS1
Nucleotide Sequence:
ATGGTGACGAAGCCGTCACATAACTTAAGAAGGGAGCACAAATGGTTAAAGGAAACGGCG ACTTTACAGGAAGACAAAGATTTCGTATTCCAGGCTATCCAAAAGCACATCGCGAACAAA AGGCCTAAGACAAATTCGCCACCAACAACGCCATCCAAAGATGAATGTGGACCAGGAACA ACAAACTTTATAACCAGCATTCCTGCATCTGGACCAACGAATACTGCTACGAAACAACAT GAAGTCATGCAAACTTTGTCGAACGATACAGAATGGCTCTCGTACACTGCCACATCGAAT CAATATGCTGACGTACCCATGGTTGATATACCAGCCAGCACAAGTGTTGTATCGAACCCG AGGACGCCCAATGGCTCGAAAACTCACAATTTCAATACTTTTCGACCGCACATGGCTAGT TCTCTTGTCGAAAATGACAGTAGCAGAAATCTAGGTAGTAGAAATAACAATAAAAGCGTA ATCGATAATTCAAGTATAGGCAAACAGCTCGAAAATGATATAAAATTAGAGGTGATCCGC CTTCAAGGCTCCCTAATTATGGCATTGAAAGAGCAATCGAAATTATTGCTACAGAAGTGT AGTATTATCGAATCCACATCATTGTCAGAGGACGCTAAAAGGCTACAGTTAAGTAGGGAT ATACGGCCTCAGTTATCAAATATGTCGATACGAATAGATAGTCTGGAAAAGGAGATCATA AAAGCCAAAAAAGATGGAATGTCAAAAGATCAAAGTAAAGGTCGCAGTCAAGTTTCTTCA CAAGATGATAATATCATATCAAGCATTCTGCCCAGCCCCTTGGAATACAATACATCTTCC AGGAATTCAAACCTGACAAGTACTACTGCGACTACCGTCACTAAGGCATTAGCCATCACT GGTGCGAAGCAAAATATTACTAATAACACCGGTAAGAATAGCAATAATGATAGTAATAAC GATGATTTGATTCAAGTCCTGGATGATGAGGATGATATAGACTGTGACCCTCCTGTAATA TTGAAGGAGGGGGCTCCACATTCTCCCGCATTCCCGCATCTCCACATGACGTCAGAAGAA CAAGATGAACTTACAAGAAGGAGAAATATGCGTTCAAGAGAACCAGTCAACTACAGAATA CCTGATAGGGATGATCCTTTTGACTATGTTATGGGTAAATCTTTGAGGGACGATTATCCG GATGTCGAAAGAGAAGAAGATGAGTTAACAATGGAGGCAGAAGATGATGCCCATTCCAGC TACATGACTACTAGAGATGAAGAAAAAGAAGAAAACGAATTACTAAATCAAAGCGATTTT GATTTTGTGGTAAACGACGACCTAGACCCAACTCAAGACACAGATTATCATGATAATATG GATGTTAGTGCAAATATTCAGGAAAGTTCTCAAGAAGGTGACACTAGGTCCACAATTACC TTGTCGCAAAATAAAAATGTTCAAGTTATTTTATCATCTCCCACAGCACAGAGCGTTCCC TCAAATGGCCAAAATCAAATAGGCGTGGAGCATATTGACTTGTTGGAAGATGATCTGGAA AAGGACGCAATTTTGGATGATAGCATGAGTTTCTCCTTTGGCCGTCAACACATGCCCATG TCTCATTCTGATCTAGAGTTGATAGACAGCGAAAAAGAAAATGAGGATTTTGAAGAAGAT AATAACAATAACGGTATCGAGTACCTATCAGATAGCGATTTAGAAAGATTTGACGAAGAA AGAGAGAATAGAACCCAAGTAGCAGATATCCAGGAACTAGACAATGACCTGAAAATAATA ACAGAAAGGAAGCTTACAGGTGACAATGAACACCCACCACCATCTTGGTCTCCCAAAATA AAAAGGGAGAAATCCAGTGTTAGTCAAAAGGATGAGGAAGACGATTTTGATGACGATTTT TCATTAAGTGATATAGTGAGTAAATCCAATTTATCTTCTAAGACGAATGGTCCAACCTAT CCTTGGTCTGATGAAGTTTTATATCGTTTACATGAAGTCTTTAAACTGCCTGGTTTTAGA CCTAACCAACTAGAGGCTGTAAATGCAACTTTGCAAGGTAAGGATGTTTTTGTTCTTATG CCAACAGGGGGTGGTAAATCTCTTTGCTATCAACTTCCTGCAGTGGTGAAATCGGGTAAA ACACATGGTACTACTATTGTCATCTCTCCGCTAATTTCCCTGATGCAAGATCAAGTGGAA CATTTATTGAATAAAAATATTAAGGCGAGCATGTTCAGTTCGAGGGGTACTGCCGAGCAA AGACGACAAACTTTCAATTTATTTATTAATGGATTATTGGATTTAGTTTACATATCTCCT GAGATGATCAGTGCCTCAGAACAATGCAAGAGAGCTATCAGTAGATTATACGCAGACGGT AAGTTGGCTCGTATTGTTGTAGATGAAGCACATTGTGTTTCTAACTGGGGCCACGATTTC AGGCCTGATTATAAAGAATTAAAATTTTTCAAAAGAGAATACCCTGATATTCCAATGATT GCTTTAACTGCAACTGCAAGTGAACAAGTCAGAATGGACATCATTCACAATTTAGAACTA AAGGAACCTGTTTTCCTAAAACAAAGTTTTAATAGAACAAATTTGTATTACGAAGTAAAC AAGAAGACCAAAAATACCATTTTTGAAATCTGTGATGCGGTTAAATCTAGGTTCAAAAAT CAAACGGGTATAATATATTGCCACTCCAAGAAATCATGCGAGCAAACATCAGCCCAAATG CAAAGAAATGGCATCAAGTGTGCATATTACCATGCAGGCATGGAGCCTGATGAAAGATTA AGTGTACAGAAGGCATGGCAGGCGGATGAGATACAAGTCATTTGTGCTACTGTTGCTTTC GGAATGGGTATTGATAAACCTGATGTGAGATTTGTTTACCACTTTACCGTTCCCCGAACA TTAGAAGGCTATTATCAAGAAACCGGCCGTGCTGGAAGAGATGGGAACTATTCATATTGT ATTACCTACTTTTCATTCAGGGACATTAGAACCATGCAGACAATGATCCAGAAGGATAAG AACTTAGACAGAGAGAATAAGGAAAAACATCTGAATAAATTACAGCAAGTAATGGCATAC TGTGACAACGTTACTGACTGCAGAAGAAAGTTAGTTTTATCTTATTTCAATGAGGATTTT GACTCCAAACTGTGTCATAAAAACTGTGATAATTGTAGAAATAGCGCCAACGTGATAAAC GAGGAAAGGGATGTTACAGAACCTGCCAAGAAGATTGTAAAATTAGTGGAAAGTATCCAA AATGAAAGAGTCACAATAATTTATTGCCAAGATGTCTTTAAAGGTTCGAGAAGCTCCAAA ATTGTTCAGGCTAACCATGACACCTTAGAGGAGCATGGTATTGGTAAATCCATGCAAAAA TCAGAAATCGAAAGGATTTTCTTCCATTTGATTACGATCCGAGTTTTACAAGAGTATTCA ATAATGAACAATAGCGGTTTTGCTTCAAGCTATGTGAAAGTTGGTCCCAATGCTAAGAAA TTGCTGACTGGAAAAATGGAGATAAAGATGCAATTTACAATATCAGCACCAAACTCCCGT CCCTCGACATCTTCTAGCTTTCAAGCAAATGAAGATAATATACCAGTTATCGCGCAGAAA TCAACAACCATTGGAGGAAATGTGGCTGCTAATCCACCGCGCTTTATAAGTGCGAAGGAA CACCTAAGATCGTATACATACGGCGGTTCGACCATGGGTAGCTCACATCCGATCACTTTG AAAAACACAAGCGATTTACGCTCGACACAAGAACTTAATAATCTGCGAATGACATACGAA CGTCTGAGGGAATTATCTTTAAATTTAGGAAATAGAATGGTTCCTCCAGTTGGGAACTTT ATGCCTGACAGTATTTTAAAAAAGATGGCAGCAATATTACCAATGAATGATTCGGCTTTT GCAACTTTAGGCACAGTGGAGGACAAATATCGCCGAAGGTTCAAGTACTTTAAAGCCACG ATAGCAGATCTTAGCAAAAAGAGATCAAGCGAGGATCATGAAAAATACGACACAATACTA AATGATGAATTTGTGAACAGGGCCGCGGCTTCCAGCAATGGGATTGCCCAAAGTACTGGA ACAAAATCTAAGTTTTTTGGCGCCAATTTAAACGAAGCCAAAGAAAATGAACAAATTATC AATCAAATCAGACAAAGCCAATTACCCAAAAATACCACAAGCAGCAAATCAGGTACAAGG TCCATCAGTAAGTCGTCCAAGAAGTCTGCTAATGGGAGACGAGGTTTTAGAAATTACCGAGGTCACTACAGAGGAAGAAAGTGA
Molecular Functions:
SGS1 is involved in ATP-dependent DNA helicase activity. Basically, SGS1 catalyzes the reaction in which water breaks down ATP to form ADP and a phosphate group. This reaction drives the unwinding of the DNA helix so that DNA replication and mRNA synthesis can occur. It is important to note, however, that nine other yeast genes have been implicated in the ATP-dependent unwinding of the DNA double helix as well. To find more information on the other genes or to learn more about SGS1's molecular function, see SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=4003.
SGS1 is necessary for the maintenance of genome stability and is essential for preserving the integrity of rDNA (ribosomal DNA) repeats. SGS1's functions in DNA replication and RNA polymerase I transcription might contribute to the phenotypes observed in Bloom's and Werner's Syndromes. (http://www.helicase.net/dexhd/SGS1.htm)
SGS1 also interacts with topoisomerase II and TOP3. "The TOP3-SGS1 protein complex may function as a eukaryotic reverse gyrase, introducing positive supercoils into extrachromosomal ribosomal DNA rings," according to Swiss-Prot. (http://us.expasy.org/cgi-bin/niceprot.pl?SGS1_YEAST.)
Biological Functions:
SGS1 is implicated in four separate biological pathways:
1. SGS1 acts to "melt" or break the interchain hydrogen bonds between double-stranded DNA in order to create two unpaired template DNA strands. This process is also known as duplex DNA melting, and eleven other yeast genes have been implicated in this phenomenon (SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=6268.)
2. Chromosome organization and biogenesis is another important function of SGS1. This includes the assembly and arrangement of the eukaryotic chromosomes, but is not restricted to the eukaryotes. (SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=7001.)
3. After metaphase I, SGS1 aids in meiotic chromosome segregation, which is the movement of chromosomes into daughter cells during meiotic cell division. (SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=45132.)
4. In mitosis, SGS1 plays a role in mitotic sister chromatid segregation. During this process, replicated chromosomes split and migrate into daughter cells. (SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=70.)
For very specific information concerning SGS1's biochemical pathways, see http://www.helicase.net/dexhd/SGS1.htm.
Scientists at MIT believe that SGS1 could be the key to a "universal aging mechanism." A gene called WRN, which is similar to SGS1, is found in humans. When WRN is mutated, it leads to Werner's Syndrome, which is characterized by rapid aging. Researchers predict that a similar mutation in SGS1 might allow further study of the life span of yeast. (http://web.mit.edu/newsoffice/tt/1997/sep10/aging.html.)
SGS1 has been found to suppress the slow-growth caused when the top3 gene is mutated. (SGD 2004 http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SGS1.)
Cellular Component:
SGS1 has been detected in the nucleolus, which is a small, dense region of the nucleus in eukaryotic cells. The nucleolus is "rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis." (SGD 2004 http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=5730.)
Homologues:
The SGS1 sequence is partially conserved in other eukaryotes, such as S. pombe (hus2), E. gossypii (AFL159W), and A. thaliana (At3g05740). Its conserved domains are the Helicase superfamily c-terminal domain, the DEXH-box helicases, and the Helicase and RNase D C-terminal. (See NCBI's Homologene for more information: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=homologene&dopt=HomoloGene&list_uids=40000.) There are structural similarities between SGS1 and the human proteins BLM and WRN. (http://www.helicase.net/dexhd/SGS1.htm.)
Amino Acid Sequence:
mvtkpshnlr rehkwlketa tlqedkdfvf qaiqkhiank rpktnspptt pskdecgpgt tnfitsipas gptntatkqh evmqtlsndt ewlsytatsn qyadvpmvdi pastsvvsnp rtpngskthn fntfrphmas slvendssrn lgsrnnnksv idnssigkql endiklevir lqgslimalk eqsklllqkc siiestslse dakrlqlsrd irpqlsnmsi ridslekeii kakkdgmskd qskgrsqvss qddniissil pspleyntss rnsnltstta ttvtkalait gakqnitnnt gknsnndsnn ddliqvldde ddidcdppvi lkegaphspa fphlhmtsee qdeltrrrnm rsrepvnyri pdrddpfdyv mgkslrddyp dvereedelt meaeddahss ymttrdeeke enellnqsdf dfvvnddldp tqdtdyhdnm dvsaniqess qegdtrstit lsqnknvqvi lssptaqsvp sngqnqigve hidlleddle kdailddsms fsfgrqhmpm shsdlelids ekenedfeed nnnngieyls dsdlerfdee renrtqvadi qeldndlkii terkltgdne hpppswspki krekssvsqk deeddfdddf slsdivsksn lssktngpty pwsdevlyrl hevfklpgfr pnqleavnat lqgkdvfvlm ptgggkslcy qlpavvksgk thgttivisp lislmqdqve hllnknikas mfssrgtaeq rrqtfnlfin glldlvyisp emisaseqck raisrlyadg klarivvdea hcvsnwghdf rpdykelkff kreypdipmi altataseqv rmdiihnlel kepvflkqsf nrtnlyyevn kktkntifei cdavksrfkn qtgiiychsk ksceqtsaqm qrngikcayy hagmepderl svqkawqade iqvicatvaf gmgidkpdvr fvyhftvprt legyyqetgr agrdgnysyc ityfsfrdir tmqtmiqkdk nldrenkekh lnklqqvmay cdnvtdcrrk lvlsyfnedf dsklchkncd ncrnsanvin eerdvtepak kivklvesiq nervtiiycq dvfkgsrssk ivqanhdtle ehgigksmqk seieriffhl itirvlqeys imnnsgfass yvkvgpnakk lltgkmeikm qftisapnsr pstsssfqan ednipviaqk sttiggnvaa npprfisake hlrsytyggs tmgsshpitl kntsdlrstq elnnlrmtye rlrelslnlg nrmvppvgnf mpdsilkkma ailpmndsaf atlgtvedky rrrfkyfkat iadlskkrss edhekydtil ndefvnraaa ssngiaqstg tkskffganl neakeneqii nqirqsqlpk nttssksgtr sisksskksa ngrrgfrnyr ghyrgrk
Protein shape:
A portion of the three-dimensional conformation for the SGS1 protein has been determined by scientists. The total protein sequence contains 1447 amino acids, but the structure in Protein Data Bank for SGS1 (listed as 1D8B) is only 81 amino acids long. According to NCBI's BLAST2, this sequence matches perfectly with a portion of the total protein sequence. The structure shown in Fig. 2 below is the projected biological model for the SGS1 protein from amino acid 1271-1351.
Amino acid sequence of structure: ELNNLRMTYERLRELSLNLGNRMVPPVGNFMPDSILKKMAAILPMNDSAFATLGTVEDKY RRRFKYFKATIADLSKKRSSE.
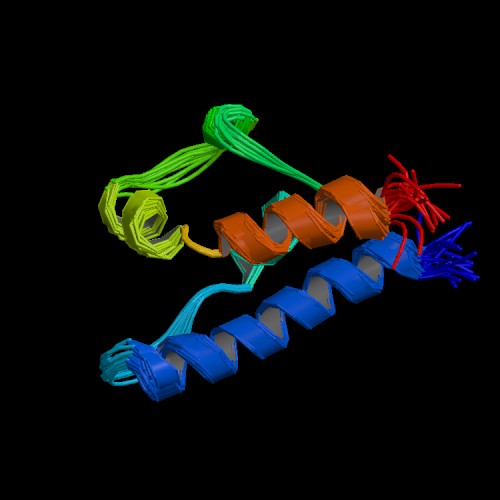
Figure 2: Assumed biological model of SGS1 protein. PDB 2004. http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics;pdbId=1D8B;page=;pid=293761097195976&bio=1&opt=show&size=500
To see the Chime file for this protein, click here. Don't have Chime? Click here to download a free copy of the Chime plug-in.
Mutants and allelic variants:
Clearly SGS1 is a requisite protein because the SGS1 yeast null mutant is inviable, according to SGD 2004. It has severe growth defects on YPD after 20 and 60 generations, and also when grown on minimal media, NaCl, or lactate. (http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SGS1.)
As mentioned above, SGS1 is structurally similar to genes that, when mutated, lead to Werner's and Bloom's Syndromes. Werner's Syndrome is characterized by a "fast-forward" aging process (see Fig. 3 below). Symptoms include graying and loss of hair, osteoporosis, cataracts, atherosclerosis, loss of skin elasticity, type II diabetes, and a propensity for certain cancers. Bloom's Syndrome causes growth deficiencies, sun-sensitivity, hypo- and hyperpigmented skin, predisposition to malignancy, and chromosomal instability. (http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=210900, http://web.mit.edu/newsoffice/tt/1997/sep10/aging.html, and http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=277700.)
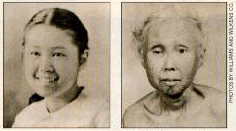
Figure 3: At age 15 (left), this Japanese American female seemed to be aging normally. By age 48 (right), however, Werner's Syndrome has rapidly aged her. NCBI 2004 http://www.ncbi.nlm.nih.gov/disease/Werner.html.
Dr. David Sinclair from Harvard Medical School has been studying the connection between Werner's Syndrome and yeast genes. "...[His] findings about SGS1 raise the possibility that the human WRN helicase is an "ALT" [alternative lengthening of telomeres] factor, and that it could be the missing link between premature aging and early onset of cancer in WS [Werner's Syndrome] patients." (http://www.hms.harvard.edu/armenise/symposia/symp5_2001/symp5_2001_day1.html)
After some experimentation, another scientist at Harvard concluded that "...mutating SGS1 caused normal aging to occur at an accelerated pace." (http://web.mit.edu/newsoffice/tt/1997/sep10/aging.html.) Harvard Professor Guarente said:
"The finding raises the possibility of locating a general aging mechanism. That would be important because we can study the aging process in a simple organism like yeast in order to learn general principles of aging..."
Yeast strains which lack SGS1 have been shown to have elevated levels of chromosome misseggregation during both mitotic and meiotic division. (SGD 2004 http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?locus=YMR190C.)
SGS1 does not necessarily implicate longevity in S. cerevisiae, but it seems as though it could prevent premature aging. Perhaps with more research, a pharmacogenomic drug based upon the activity of SGS1 could be used to effectively reverse the Werner's Syndrome phenotype.
YMR187C
Nucleotide Sequence:
ATGACCCCACCTCATTTTTTTTTATCTCTCATAAAAAAAAGATGCATATGCTGGATATGTTTGGAAGAATCCACATATGACTCTACATGGCTACAGCACACGTGCGGTTGCAACTTACAA ATTCATAAACGCTGCTATATAAGATGGCTCTATCAAATGCATGTTGAACTTTTTCTTCCGAACACAGTGGACCTACCAAAGGATGCTGATTTACCTATAATCACCTGTTTAAAATGTCTC GTTGATGGGCACCATGATTTTATGACGACTTTCTCTCTTACAGAAATATGGGAAACTCGTCCCATTTGGGGGCAAAAATCAGTACCGTTTCAAAATGACTACGTGTTCAATCTAATGTCT CTTTATACCAAACGGGATAATCATCCACCTTATGTGTTAGTGAAATTTGGCGAATGTCCTCAATGCAAGAAAACAAATTTCATAAAAAGACCTACAGTAACCATCCAGTCTTCTGTACTA TCCCTGTTTTATCAGTGGCAAAAAATTACACGATACGTGATACCATTAGGGATAACTAGTTTATTTCTTTTAAACCCTGAAAAGACAAGTTTTGACATTGGATTATGGCAACTACGCTGC CTTTTCCCTGAGAATGTTCTACGAAATATGTTAAACATCAGCACTACAAAGGCATTGGACGTATATGCGCAAACTGAACGTGGCTTATTAAGCATTCCTTTAACCTCCTCGATCATAATA TATGGGTTCATACATTATCTTAGCAACATTTCTAACGTCAGTGCTAATGCAATACTCTTTAAGTGGGTGTATCTCAGTATTGTAAAAACTGCTGGAAACAAATATTACAAGGGTATAGGT TTACCTAAAATAATTTTGTATTCTAACTTGGCTACATTTTGCTATAATTTTACCTTTAAA AGATTGGTTGACTTAATTTACAGGAGGCTGATCAATAAAGGAGGTAAGTATTTGTATCAC GGAAATTTTGAAAACTCTAGTAATAGCGTTCCAGCTGAGGAATTTTTTATCAGGCGGAATTGGTATGCTATTCTTGCAGAAAAGATACTATGGCCCTTTGTTGGTAAATGTACAGGAGGT CTTCTATTGAATGCTTTTCTATGGATACAGAGAAAGTTTAAAATTGAATGGACGCCGAATTGCTCCCCTTCGGAATTTAGAATGATTTTCAATATTATAGGTTGTGGTACTGCTGCTATA GGCTGGTCAAGTCTGAAGCTCTATGCCAGTTACAAAAGATGTCAGGAACTTGAAAAGATCAATGAATTTATTGAACAAAGTTGCAAAGGTGAGTAA
BLASTn:
After performing a BLASTn on the YMR187C nucleotide sequence, it seems as though there are two feasible possibilities for this open reading frame. Results shown in Fig. 4:
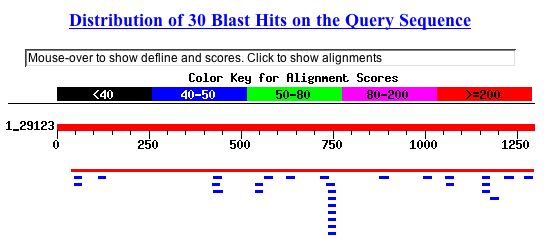

Figure 4: BLASTn results from YMR187C nucleotide sequence. NCBI BLAST 2004 http://www.ncbi.nlm.nih.gov/blast/.
First, it could be an inhibitory protein that prevents the expression of SGS1. The sequence had a 0.8 expect value and 100% identity with a predicted gene in gallus gallus (chicken). This gene predicted to occur in chickens might parallel the secreted Xwnt8 inhibitor "sizzled," which is found in Xenopus laevis. Xwnt8 is implicated in anteroposterior patterning, according to NCBI.
Next, the sequence could be related to a macaca mulatta (rhesus monkey) major histocompatibility complex gene. This sequence had a 0.8 expect value, as well, and also had 100% identity. There is a human histocompatibility complex gene similar to the monkey gene. The integral protein functions in immune response. (NCBI 2004 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=Graphics&list_uids=3135.)
Amino Acid Sequence:
Blastx and NCBI Protein both provided the same predicted amino acid sequence, which is shown below:
mtpphfflsl ikkrcicwic leestydstw lqhtcgcnlq ihkrcyirwl yqmhvelflp ntvdlpkdad lpiitclkcl vdghhdfmtt fslteiwetr piwgqksvpf qndyvfnlms lytkrdnhpp yvlvkfgecp qckktnfikr ptvtiqssvl slfyqwqkit ryviplgits lfllnpekts fdiglwqlrc lfpenvlrnm lnisttkald vyaqtergll sipltssiii ygfihylsni snvsanailf kwvylsivkt agnkyykgig lpkiilysnl atfcynftfk rlvdliyrrl inkggkylyh gnfenssnsv paeeffirrn wyailaekil wpfvgkctgg lllnaflwiq rkfkiewtpn cspsefrmif niigcgtaai gwsslklyas ykrcqeleki nefieqsckg e
Other Observations:
NCBI's Conserved Domain Search found no hits for the protein sequence. (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.)
HomoloGene returned no hits for YMR187C. (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=Search&DB=homologene.)
The Kyte-Doolittle hydropathy plot indicates five transmembrane domains in Fig. 5.
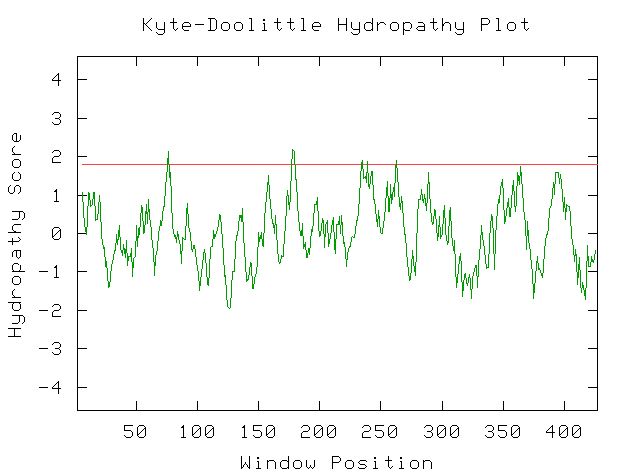
Figure 5: Kyte-Doolittle hydropathy plot for YMR187C sequence. One transmembrane domain is indicated. 2004 http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm.
From this information, it seems as though YMR187C might code for an integral protein.
A PREDATOR search of amino acid sequence returned the following information:

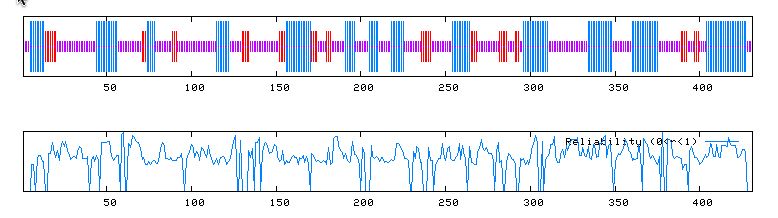
Figure 6: PREDATOR prediction of YMR187C protein conformation from its amino acid sequence. PREDATOR 2004 http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl.
Here, it can be seen that the protein might be composed of alpha helices, many random coils, and some extended strands.
SGD 2004 reports no growth defect detected on YPD after 20 generations or on minimal media. A moderate growth defect occurs on YPD after 60 generations or on lactate. (SGD 2004 http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YMR187C.)
YMR187C is located just upstream from a gene called MRPS17 which is a mitochondrial small ribosomal subunit that acts as a structural component of the ribosome and participates in aerobic respiration and protein biosynthesis. (SGD 2004 http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YMR188C.)
Conclusion:
From the information I obtained about YMR187C, a few options for the function of this non-annotated yeast gene exist. Perhaps it could code for a protein that inhibits the effects of SGS1, as its sequence is slightly similar to the predicted g. gallus protein, which acts like the Xwnt8 inhibitor Sizzled in Xenopus. But this choice does not seem logical because SGS1 is so important for genome stability and viability of an organism. SGS1 is also important for lifelong function, not just developmental processes.
YMR187C might function like the gene found in monkeys for the histocompatibility complex. Since there were five transmembrane domains found with the Kyte-Doolittle hydropathy plot, it seems plausible.
Maybe YMR187C codes for a protein involved in some sort of DNA process like mitosis, meiosis, replication, or transcription. It lies on chromosome XIII near SGS1, which plays important roles in DNA unwinding and chromosome segregation; and it is also near MRPS17, which is a structural component of mitochondrial ribosomes. It seems as though this region of S. cerevisiae in particular codes for proteins vital to DNA processes.
The final option is that this particular ORF is, in fact, not a coding portion of DNA and lies in an intergenic region.
References:
[COG] Clusters of Orthologous Groups. 2004 Oct 7. COG0514: RecQ Superfamily II DNA helicase. http://www.ncbi.nlm.nih.gov/COG/new/release/cow.cgi?cog=COG0514. Accessed 2004 Oct 7.
[NCBI] National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/ Accessed 2004 Oct 7.
[PDB] Protein Data Bank. 1D8B. http://www.rcsb.org/pdb/cgi/explore.cgi?pid=203151097204076&pdbId=1D8B. Accessed 2004 Oct 7.
[PREDATOR] Network Protein Sequence Analysis. http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html. Accessed 2004 Oct 7.
[SGD] Saccharomyces Genome Database. 2004 Oct 7. SGS1/YMR190C. http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YMR190C. Accessed 2004 Oct 7.
[SP] Swiss-Prot. P35187. http://us.expasy.org/cgi-bin/niceprot.pl?SGS1_YEAST. Accessed 2004 Oct 7.
Bennett RJ, Keck JL. 2004. Structure and Function of RecQ DNA Helicases. Critical Reviews in Biochemistry and Molecular Biology, 39(2): 79-97. See abstract at the following link: http://www.crbmb.com/cgi/content/abstract/39/2/79. Accessed 2004 Oct 7.
Sinclair D. 2001Giovanni Armenise-Harvard Foundation 5th Annual Symposium. RecQ helicases provide links between premature aging and cancer. See http://www.hms.harvard.edu/armenise/symposia/symp5_2001/symp5_2001_day1.html for summary and http://www.hms.harvard.edu/pathol/sinclair/Pages/recqfamily.html for RecQ DNA helicase family graphic. Accessed 2004 Oct 7.
Wright SH. 1997 Sep 10. Gene with role in yeast life span discovered. MIT News Office. http://web.mit.edu/newsoffice/tt/1997/sep10/aging.html. Accessed 2004 Oct 7.
http://www.helicase.net/dexhd/SGS1.htm. SGS1. Accessed 2004 Oct 7.
____________________________________________________________________________________________________________________________________________________
For more information, please refer to the Genomics Home Page or to the Davidson College Home Page.
Questions or comments? E-mail me: "jehoekstra" at "davidson.edu".
Back to my genomics home page.
Page last updated 2004 Oct 8 by J. Hoekstra.