Expression of ADY2 and YCR007C
ADY2
Quick Background:
Molecular Function: ADY2 codes for the transmigrate Ady2p protein which directs the active transport of several macromolecules, small molecules, and ions across, between or within cells.
Biological Process: This gene has several biological processes. First, it plays a major role in DNA metabolism, involved in the accumulation of DYads, and therefore important in meiosis. ADY2 is also involved in acetate transport. A third biological process that this gene contributes to is nitrogen utilization and ammonia production. Ammonia is important as a starvation signal between yeast colonies, as discussed by Palkova, et al. (2002, <source>)
Cellular Component: Ady2 is a transmembrane protein and is also found in the mitochondrion.
Expression:
All of the following images are from Expression Connection (2004, <homepage>) unless otherwise stated.

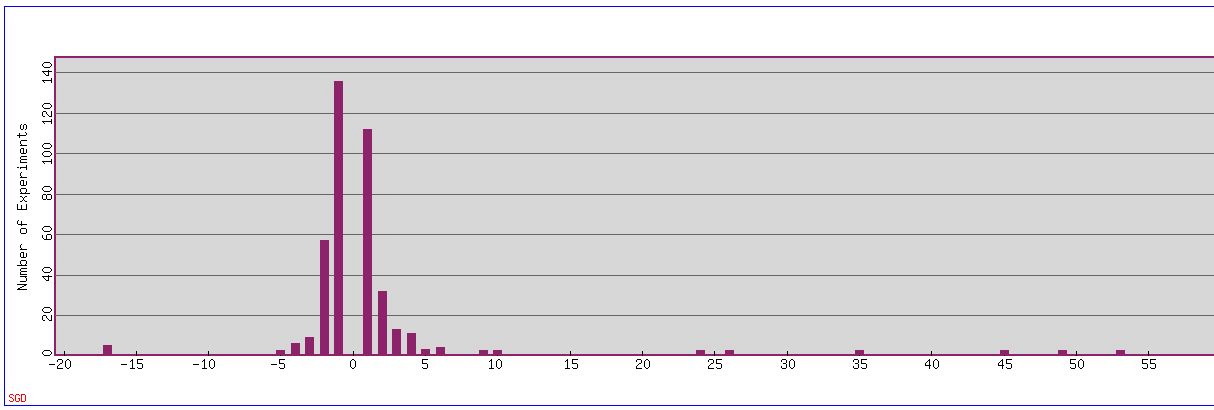
Fig. 1: Expression Histogram in summary of several different experiments for ADY2. Negative numbers signify repression, while positive signifies induction.
Sporulation (Expression during sporulation Chu S, et al.)
Below is the microarray expression for ADY2 in the at different points in the sporulation process. In this experiment, the max induction was 5.9, and the max repression was -1.0. As can be seen, at about .5 hours, the gene is highly induced, and continues throughout the entire sporulation event. Sporulation includes both meiosis and spore morphogenesis, and this microarray supports that meiosis is one of the biological functions of ADY2. We know that ADY2 is involved in acetate transport, which is involved in the release of energy, required for sporulation.

Fig. 2: Microarray of ADY2 in the sporulation experiment.

Fig. 3: Microarray data of several genes whose expression patterns seem to parallel those of ADY2 in this experiment. (click here)
Above we can see a chart of the genes whose expression patterns most closely matched those of ADY2. It is interesting to see that while some are specifically associated with transport as well (such as ISA1 and OAC1), most are known to be involved in different areas necessary for meiosis. Also, there does not seem to be a pattern to the location of the gene products.
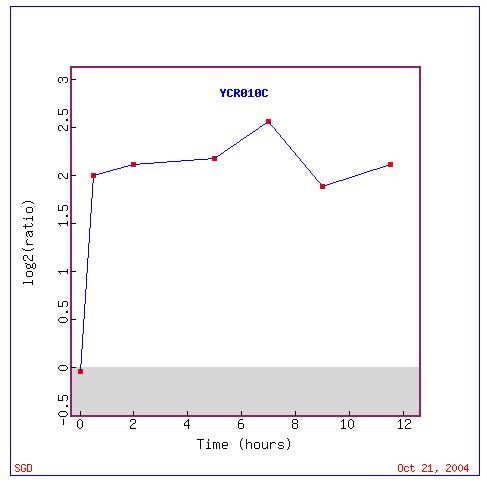
Fig. 4: Graph of ADY2 (also known as YCR010C) expression as a ratio throughout the experiment.
In the graph above, we can see in numbers the amount of induction of ADY2, reaching up to 2.5 (5.9 fold) induction close to hour 8.
Response to Alpha factor (Expression in response to alpha-factor (over time) Roberts CJ, et al.)

Fig. 5: Microarray of ADY2 in alpha factor over time experiment.
Alpha factor is a pheromone used by yeast to indicate the presence of the opposite mating type, which leads to preparation for sexual reproduction. This experiment showed that the most highly induced genes under these conditions were those associated with the activation and regulation of signal transportation/transduction. Since ADY2 is involved in molecular transport, it makes sense that it would be highly induced under these conditions. In this experiment, the max induction was 6.1 and the max repression was -1.3.
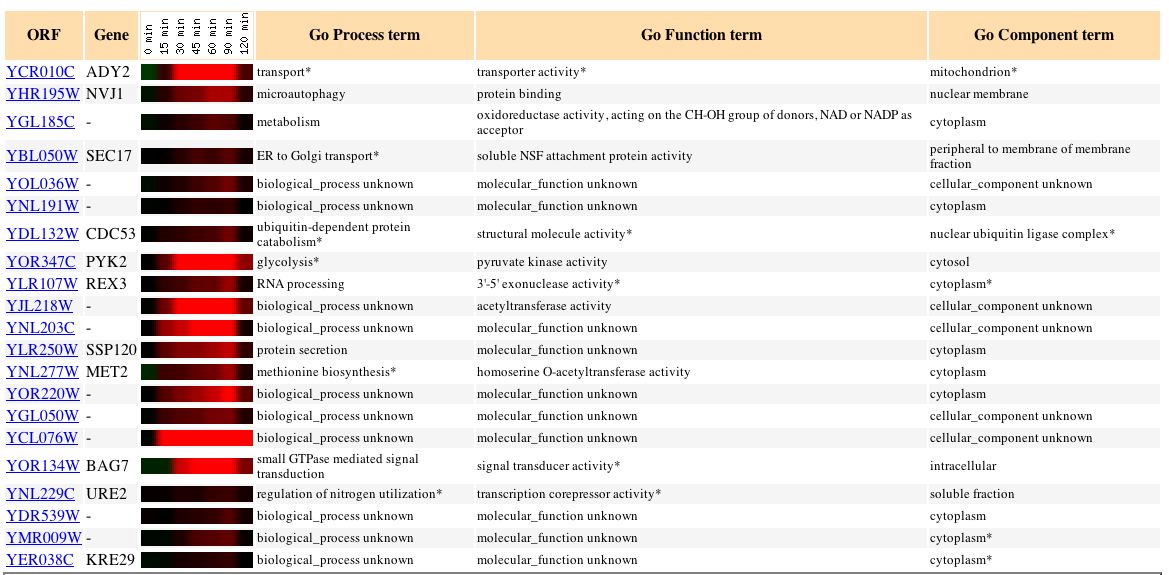
Fig. 6: Microarray data of several genes whose expression patterns seem to parallel those of ADY2 in this experiment. (original)
Above we see several genes that have a similar expression pattern to ADY2 in this experiment. A large number of their biological processes are unknown, or not fully described. Below we see ADY2 expression in this experiment shown in graph form over time. We can see that ADY2 has above a 2.5 (6.9 fold) induction at around 90 minutes.
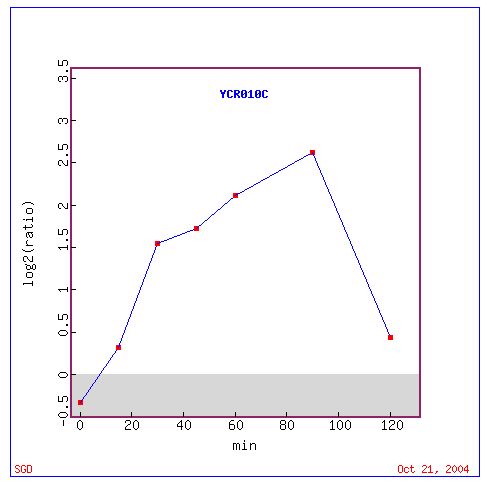
Fig. 7: Graph of ADY2 (also known as YCR010C) expression as a ratio throughout the experiment.
We can compare these results to those of the effect of different alpha factor concentrations. The microarray for this experiment is shown below. (Expression in response to alpha-factor (various concentrations) Roberts CJ, et al., <homepage >) In this experiment, the max induction was 3.8 and max repression was -1.1.

Fig. 8: Microarray of ADY2 in alpha factor concentration experiment.
YCR007C
YCR007C is a suspected gene whose biological process, molecular function, and cellular component are unknown. In a previous assignment I hypothesized that since it belongs to the DUP240 gene family, it is likely to have similar functions. The two known genes in this family have as their biological function protein binding, the process they accomplish is vesicle organization, and their cellular components are the golgi apparatus, the endoplasmic reticulum, and the plasma membrane.
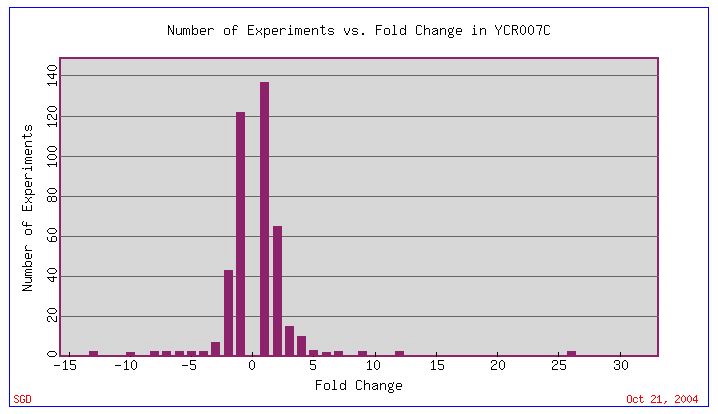
Fig. 9: Expression Histogram in summary of several different experiments for YCR007C. Negative numbers signify repression, while positive signifies induction. (original)
Expression during sporulation (Chu S, et al.)

Fig. 10: Microarray of YCR007C in the sporulation experiment.
In the above figure, we can see that this gene is highly repressed in after time .5 hours of this experiment. Here, the max repression is -3.4, and the max induction was not significant. This gene seems most highly repressed between time .5 hours, and time 7 hours, the time during sporulation at which meiosis I and II are taking place.
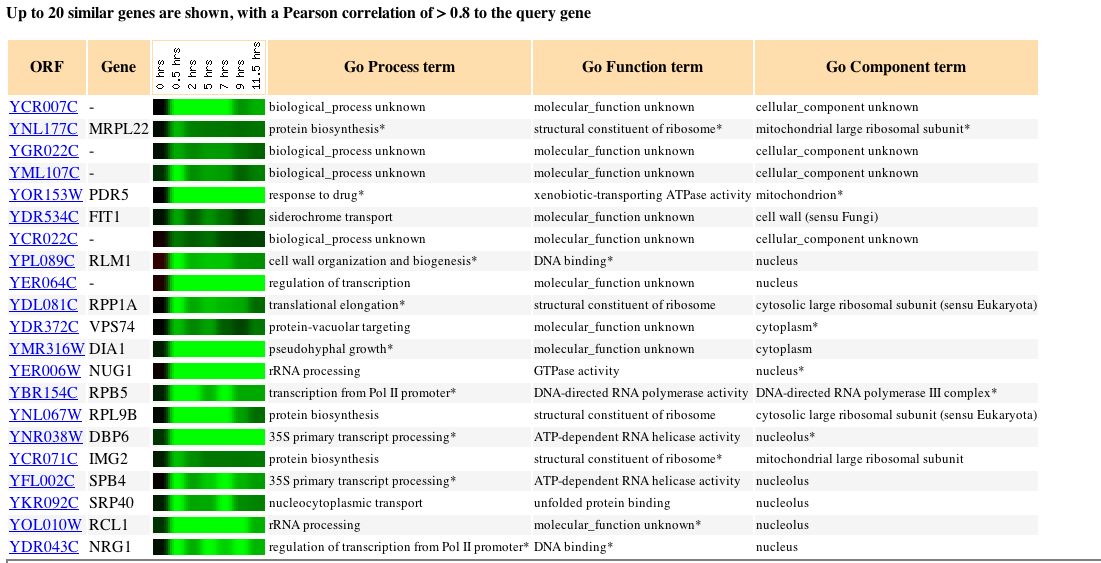
Fig. 11: Microarray data of several genes whose expression patterns seem to parallel those of YCR007C in this experiment. (original)
As can be seen from the figure above, many of the genes that are regulated similarly to YCR007C have either unknown or uncertain functions. Many of these genes are associated with protein biosynthesis and translation. If we take the "guilt by association" approach, we could assume that the gene product of YCR007C is involved in some form of protein binding.
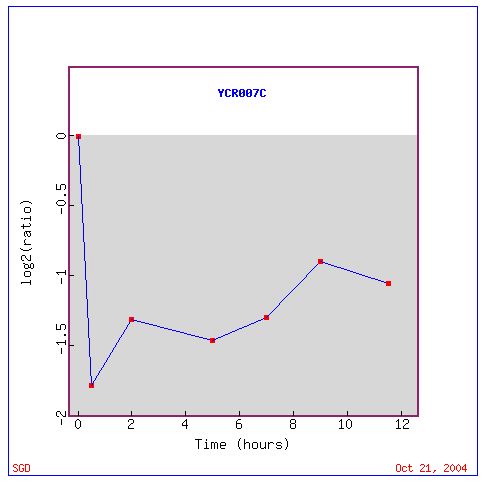
Fig. 12: Graph of YCR007C expression as a ratio throughout the experiment.
Above we see a graph of the expression of YCR007C throughout the experiment. It shows that this gene was most highly repressed early on in the sporulation process and slackened off towards the end.
Alpha factor over time (Expression in response to alpha-factor (over time) Roberts CJ, et al. )
As seen in ADY2, this experiment tests gene expression in response to the pheromone alpha factor. In this experiment, the maximum induction of YCR007C is 2.9. Below we see a picture of its microarray, where it is slowly induced starting at 15 minutes and increases until the end of the experiment.

Fig. 13: Microarray of YCR007C in alpha factor experiment.
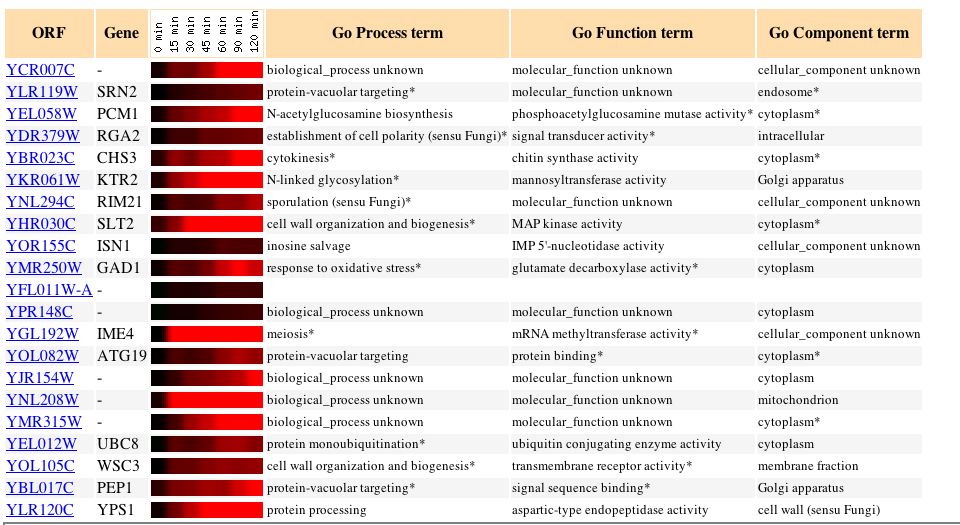
Fig. 14: Microarray data of several genes whose expression patterns seem to parallel those of YCR007C in this experiment. (original)
Knowing that alpha factor stimulates sexual reproduction in yeast, it would make sense that genes involving protein processing in preparation for this event would be induced. As can be seen by the figure above, many of the similarly expressed genes are involved in protein-vacuolar targeting, which is similar to the function predicted for YCR007C.
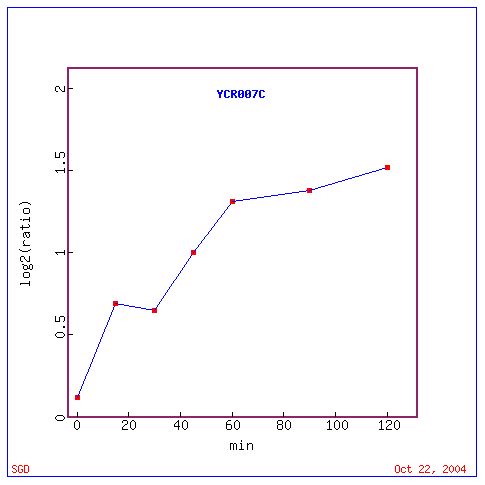
Fig. 15: Graph of YCR007C expression as a ratio throughout the experiment.
Calcineurin Pathway (Expression regulated by the calcineurin/Crz1 pathway Yoshimoto H, et al.)
This experiment showed that YCR007C is a calcineurin dependent gene that is regulated by Ca2+. The max induction was 26.2 and the max repression was -13.3. Below the microarray data can be seen.

Fig. 16: Microarray of YCR007C in calcineurin experiment.
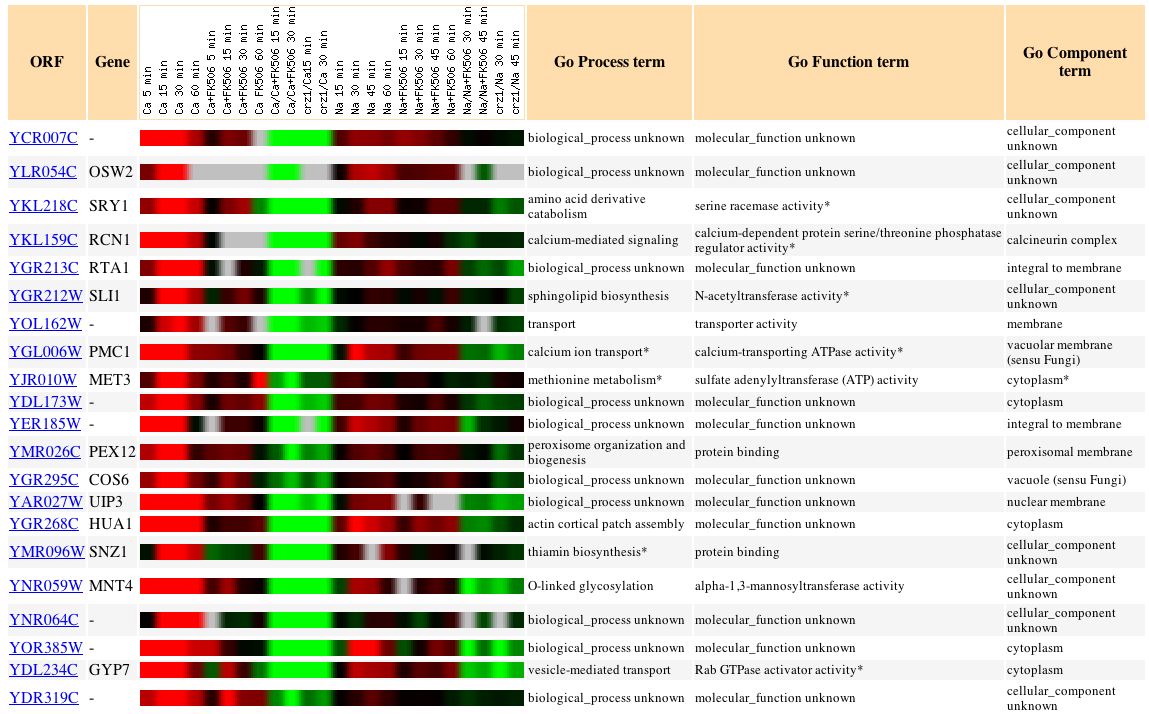
Fig. 17: Microarray data of several genes whose expression patterns seem to parallel those of YCR007C in this experiment. (original)
As can be seen from the figure above, most of the genes whose expression patterns are similar to YCR007C have unknown functions and/or processes. The conclusion that Yoshimoto, et al., come to states that YCR007C is a calcineurin dependent gene regulated by the calcium ion, not the sodium ion. According to them, calcineurin-dependent genes function in signaling pathways, ion/small molecule transport, cell wall maintenance, and vesicular transport, supporting the prediction made previously that the YCR007C gene product is involved in protein binding and vesicle organization.
Conclusion
While we cannot be sure of the function of YCR007C, using the "guilt by association" technique, we can attribute its function to protein binding and vesicle organization, originally predicted from the gene family.
Sources
All information obtained through Expression Connection (2004, <homepage>)
Davidson College Biology Department